Abstract
The silver stripped skipper, Leptalina unicolor Bremer and Grey, 1853 (Lepidoptera: Hesperiidae), is listed as endangered insect in South Korea. We sequenced the whole genome (15,854 bp) of L. unicolor species using Next-Generation Sequencing method and the subsequent gap-filling method. This genome included a set of typical genes and one major non-coding A + T-rich region, with an arrangement identical to that observed in most lepidopteran genomes. Twelve protein-coding genes (PCGs) had the typical ATN start codon, whereas COI had the atypical CGA codon that is frequently found in the start region of the lepidopteran COI. The 757-bp long A + T-rich region was the second largest among completely sequenced Hesperiidae, which ranged from 234 to 793 bp. Phylogenetic reconstruction was performed by maximum-likelihood method using the concatenated sequences of 13 PCGs and two rRNAs of available species of Hesperiidae, including that of L. unicolor (a total of 28 species). The resulting phylogeny provided strong support for monophyletic Heteropterinae in which L. unicolor belongs, with the highest nodal support and a sister relationship between current L. unicolor and co-subfamilial species Carterocephalus silvicola with a bootstrap value of 91%.
Leptalina is a monotypic temperate east Asian genus. The silver stripped skipper, Leptalina unicolor Bremer and Grey, 1853 (Lepidoptera: Hesperiidae), is distributed in Japan, eastern China, Russia Amur, and throughout Korea, except for the remote island of Jeju (Seok Citation1973). It occurs 2–3 times per year in the warmer region and once a year in the cold and mountainous region of Japan (Fukuda et al. Citation1984). In Korea, adults occur twice per year, showing seasonal dimorphism; the summer type adults produce more eggs than the spring type adults (Hong et al. Citation2016). In Korea, this species is under sudden decline in population (Ministry of Environment Citation2012) and, therefore, is listed as an Endangered species. Leptalina unicolor population decline has also been reported in Japan and Russia, and is listed as a Near Threatened species (Mano and Fujii Citation2009).
In 2016, an adult L. unicolor was collected from Inje-gun, Gangwon-do Province, South Korea (38° 01′ 39.92″ N, 128° 25 58.22″ E), and subsequently deposited as a voucher specimen at the Chonnam National University, Gwangju, Korea, under accession no. CNU7526. To sequence the mitogenome of L. unicolor, a library was prepared using the TruSeq Nano DNA Sample Preparation Kit. Whole genome sequencing was performed on the Illumina NextSeq-500 platform using a Mid Output v2 kit to produce 150 bp paired-end reads (Illumina, San Diego, CA, USA). Construction of the genome was conducted using MITObim ver. 1.9 (Hahn et al. Citation2013). Annotation of various genomic features was conducted using MITOS WebServer (http://mitos.bioinf.uni-leipzig.de/index.py) and subsequent manual BLAST search (Altschul et al. Citation1990). To fill and clarify gaps, a long fragment encompassing ND5 and lrRNA genes was amplified and then short fragments (CytB∼ND1, ND1, and ND1∼lrRNA) were individually amplified using published primer sets (Kim et al. Citation2012). Phylogenetic analysis was performed using available species in the family Hesperiidae, including L. unicolor. Thirteen protein-coding genes (PCGs) and two rRNA genes of 30 mitogenome sequences, including those of the two outgroup species, were aligned and concatenated (12,487 bp including gaps). The substitution model, GTR + GAMMA + I, was selected using Modeltest ver. 3.7 (Posada and Crandall Citation1998). The maximum-likelihood (ML) method was conducted using RAxML-HPC2 on XSEDE ver. 8.0.24 (Stamatakis Citation2014), implemented on the CIPRES Portal ver. 3.1 (Miller et al. Citation2010). Trees were visualized with FigTree ver. 1.42 (http://tree.bio.ed.ac.uk/software/figtree/).
The complete 15,854-base pair (bp) mitogenome of L. unicolor is composed of typical gene sets (two rRNAs, 22 tRNAs, and 13 PCGs) and a major non-coding A + T-rich region (GenBank acc. no. MK265705). The length of A + T-rich region of L. unicolor is 757 bp, which is the second largest in Hesperiidae (793 bp in C. silvicola, Kim et al. Citation2014). The gene arrangement of the L. unicolor mitogenome is identical to that of the ditrysian Lepidoptera (Kim et al. Citation2010). However, cofamilial Erynnis montanus has been reported to have trnS-trnN arrangement at the tRNA cluster region located between ND3 and ND5 genes, instead of trnN-trnS arrangement (Wang et al. Citation2014). Twelve PCGs had the typical ATN start codon, whereas COI gene had an atypical CGA codon frequently found in the start region of the lepidopteran COI gene (Kim et al. Citation2012).
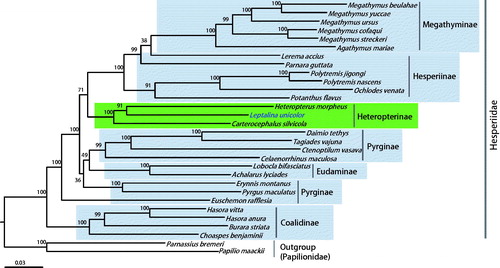
Phylogenetic analyses showed that Heteropterinae, which includes current L. unicolor, formed a strong monophyletic group (Bootstrap support [BS] = 100%). Likewise, Megathyminae (100%), Eudaminae (100%), Pyrginae (100%), and Coalidinae (100%) were strongly supported as monophyletic groups, whereas Hesperiinae and Pyrginae were non-monophyletic groups (). Lerema accius and Parnara guttata in Hesperiinae grouped together with Megathyminae, forming a strongly supported group (99%). Euschemon rafflesia in Pyrginae placed as the sister to all subfamilies, except for Coalidinaeo, whereas the other two co-subfamilial species, E. montanus and Pyrgus maculatus in Pyrginae, formed a strong monophyletic group. A previous mitogenome-based phylogeny showed that Potanthus flavus, L. accius, and P. guttata in Hesperiinae formed a strong monophyletic group (Zhang, Cong, Shen, et al. Citation2017). This result is inconsistent with the present results, indicating the need for further in-depth analysis and taxon sampling. The placement of E. rafflesia as a sister to the rest of Hesperiidae subfamilies, except for Coeliadinae, was also supported in previous mitogenome-based phylogeny (Zhang, Cong, Shen, et al. Citation2017).
Figure 1. Phylogenetic tree of Hesperiidae, using maximum-likelihood (ML) method based on concatenated 13 protein-coding genes (PCGs) and 2 rRNAs. The numbers at each node specify bootstrap percentages of 1000 pseudoreplicates. Papilionidae (Papilio maakii and Parnassius bremeri) were utilized as the outgroup. GenBank accession numbers are as follows: Pyrgus maculatus, KP689265 (unpublished); Euschemon rafflesia, KY513288 (Zhang, Cong, Shen, et al. Citation2017); Tagiades vajuna, KX865091 (Liu et al. Citation2017); Erynnis montanus, KC659955 (Wang et al. Citation2014); Celaenorrhinus maculosa, KF543077 (Wang et al. Citation2015); Daimio tethys, KJ813807 (Zuo et al. Citation2016); Ctenoptilum vasava, JF713818 (Hao et al. Citation2012); Achalarus lyciades, KX249739 (unpublished); Lobocla bifasciatus, KJ629166 (Kim et al. Citation2014); Heteropterus morpheus, KF881050 (unpublished); Carterocephalus silvicola, KJ629163 (Kim et al. Citation2014); Polytremis jigongi, KP765762 (unpublished); Parnara guttata, JX101619 (unpublished); Potanthus flavus, KJ629167 (Kim et al. Citation2014); Lerema accius, KT598278 (Cong and Grishin Citation2016); Ochlodes venata, HM243593 (unpublished); Polytremis nascens, KM981865 (Jiang et al. Citation2016); Choaspes benjaminii, JX101620 (unpublished); Hasora anura, KR189008 (unpublished); Hasora vitta, KR076553 (unpublished); Burara striata, KY524446 (Zhang, Cong, Fan, et al. Citation2017); Megathymus yuccae, KY630500 (Zhang, Cong, Fan, et al. Citation2017); M. streckeri, KY630501 (Zhang, Cong, Fan, et al. Citation2017); M. ursus, KY630502 (Zhang, Cong, Fan, et al. Citation2017); M. cofaqui, KY630503 (Zhang, Cong, Fan, et al. Citation2017); Agathymus mariae, KY630504 (Zhang, Cong, Fan, et al. Citation2017); M. beulahae, KY630505 (Zhang, Cong, Fan, et al. Citation2017); Parnassius bremeri FJ871125 (Kim et al. Citation2009); and Papilio maackii KC433408 (Dong et al. Citation2013).
Disclosure statement
No potential conflicts of interest are reported by the authors.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Cong Q, Grishin NV. 2016. The complete mitochondrial genome of Lerema accius and its phylogenetic implications. PeerJ. 4:e1546.
- Dong Y, Zhu LX, Wu YF, Wu XB. 2013. The complete mitochondrial genome of the Alpine black swallowtail. Papilio Maackii (Insecta: Lepidoptera: Papilionidae). Mitochondr DNA. 24:639–641.
- Fukuda H, Hama E, Kuzuya T. 1984. The life histories of butterflies in Japan 4. Osaka: Hoikusha (In Japanese with English summary).
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucl Acids Res. 41:e129.
- Hao J, Sun Q, Zhao H, Sun X, Gai Y, Yang Q. 2012. The complete mitochondrial genome of Ctenoptilum vasava (Lepidoptera: Hesperiidae: Pyrginae) and its phylogenetic implication. Comp Funct Genomics. 2012:1.
- Hong SH, Yoon CS, Kim HG, Cheong SW. 2016. Life cycle and breeding information of Leptalina unicolor from Korea (Lepidoptera: Hesperiidae). J Environ Sci Int. 25:1633–1641.
- Jiang W, Zhu J, Yang Q, Zhao H, Chen M, He H, Yu W. 2016. Complete mitochondrial DNA genome of Polytremis nascens (Lepidoptera: Hesperiidae). Mitochondr DNA A. 27:3131–3132.
- Kim JS, Park JS, Kim MJ, Kang PD, Kim SG, Jin BR, Han YS, Kim I. 2012. Complete nucleotide sequence and organization of the mitochondrial genome of eri-silkworm, Samia cynthia ricini (Lepidoptera: Saturniidae). J Asia Pac Entomol. 15:162–173.
- Kim MI, Baek JY, Kim MJ, Jeong HC, Kim KG, Bae CH, Han YS, Jin BR, Kim I. 2009. Complete nucleotide sequence and organization of the mitogenome of the red-spotted apollo butterfly, Parnassius bremeri (Lepidoptera: Papilionidae) and comparison with other lepidopteran insects. Mol Cells. 28:347–363.
- Kim MJ, Wan X, Kim KG, Hwang JS, Kim I. 2010. Complete nucleotide sequence and organization of the mitogenome of Endangered Eumenis autonoe (Lepidoptera: Nymphalidae). Afr J Biotechnol. 9:735–754.
- Kim MJ, Wang AR, Park JS, Kim I. 2014. Complete mitochondrial genomes of five skippers (Lepidoptera: Hesperiidae) and phylogenetic reconstruction of Lepidoptera. Gene. 549:97–112.
- Liu FF, Li YP, Jakovlić I, Yuan XQ. 2017. Tandem duplication of two tRNA genes in the mitochondrial genome of Tagiades vajuna (Lepidoptera: Hesperiidae). Eur J Entomol. 114:407–415.
- Mano T, Fujii H. 2009. Decline and conservation of butterflies and moths in Japan VI. Tokyo: Lepidopterological Society of Japan; p. 107–265.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the 9th Gateway Computing Environments Workshop (GCE), New Orleans. p. 1–8.
- Ministry of Environment (South Korea). 2012. Red data book of Endangered insects in Korea I. Sejong: Ministry of Environment (South Korea).
- Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics. 14:817–818.
- Seok JM. 1973. The distribution maps of butterflies in Korea. Seoul: Bojinjae.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang AR, Jeong HC, Han YS, Kim I. 2014. The complete mitochondrial genome of the mountainous duskywing, Erynnis montanus (Lepidoptera: Hesperiidae): a new gene arrangement in Lepidoptera. Mitochondr DNA. 25:93–94.
- Wang K, Hao J, Zhao H. 2015. Characterization of complete mitochondrial genome of the skipper butterfly, Celaenorrhinus maculosus (Lepidoptera: Hesperiidae). Mitochondr DNA. 26:690–691.
- Zhang J, Cong Q, Fan XL, Wang R, Wang M, Grishin NV. 2017. Mitogenomes of giant-skipper butterflies reveal an ancient split between deep and shallow root feeders. F1000Res. 6:222.
- Zhang J, Cong Q, Shen J, Fan XL, Wang M, Grishin NV. 2017. The complete mitogenome of Euschemon rafflesia (Lepidoptera: Hesperiidae). Mitochondr DNA B. 2:136–138.
- Zuo N, Gan S, Chen Y, Hao J. 2016. The complete mitochondrial genome of the Daimio tethys (Lepidoptera: Hesperoidea: Hesperiidae). Mitochondr DNA A. 27:1099–1100.
