Abstract
The Collared Crow (Corvus pectoralis), in the order Passeriformes, it widely distributed in large areas encompassing China and northern Vietnam. It is a vulnerable bird that is of international concern. In this study, we first sequenced and described the complete mitochondrial genome and phylogeny of C. pectoralis. The results showed that the whole genome of C. pectoralis was 16,857 bp long and contains 13 PCGs, 2 ribosomal RNA genes, 23 transfer RNA genes, and 1 loop region. The overall base composition of the mitochondrial DNA was 31.13% for A, 29.52% for C, 24.46% for T, and 14.89% for G, with a GC content of 44.41%. The phylogenetic tree showed that C. pectoralis was clustered with C. brachyrhynchos and then together with other two crows in family Passeriformes. This information will be useful in the current understanding of the phylogeny and evolution of Passeriformes.
Corvus pectoralis, which belongs to the Passeriformes, widely distributed in China and northern Vietnam (John et al. Citation2015). It can be easily found in open areas where trees are scattered, especially near waters. In recent years, due to the intensification of human agriculture and the consequent overuse of pesticides and rodenticides, the reduction in prey has led to a decline in the population of the species within its range (Londei Citation2013), and it has been listed as a vulnerable group (VU) on the IUCN Red List of Threated Species (IUCN, Citation2019). Despite this, genetic information of C. pectoralis is quite limited. Haring et al. (Citation2012) explored the appearance variation of the genus Corvus species based on the DNA sequence of the mitochondrial control region, and did not study the evolutionary classification of this species based on the complete mitochondrial genome. Therefore, it’s necessary to sequence the complete mitochondrial genome of C. pectoralis to enhance our understanding of the phylogeny and evolution of Corvidae.
The whole mitochondrial DNA was extracted from muscle specimen with DNeasy Plant Mini kit (Qiagen, Valencia, CA) and specimen was collected from Dongting Lake and stored at Hunan Engineering Research Centre for Internet of Animals, China with accession number 20170801PC. The genomic DNA data were sequenced by Illumina Miseq platform (Illumina, San Diego, CA). The adapter and low quality reads were filtered out by NGS QC toolkit (Patel and Jain Citation2012). The genome was annotated using the MITOS online service (Bernt et al. Citation2013). Annotated PCGS were compared with other vertebrate species sequences and the method referred to Zhang’s addressing (Zhang et al. Citation2017). The complete mitochondrial genome of C. pectoralis has been submitted to the NCBI database with the accession number of MN310552. Phylogenetic tree among C. pectoralis and its related orders were presented using 13 PCGs by Neighbor-joining analyses in MEGA 7.0 with 1000 bootstrap replicates (Kumar et al. Citation2016).
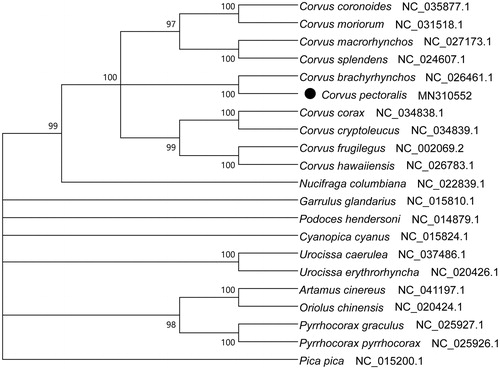
The complete mitogenome of C. pectoralis is 16 857 bp long and contains 13 PCGs, 2 ribosomal RNA genes, 23 transfer RNA genes, and 1 loop region. This feature was similar to the typical mitogenome of other birds (Ren et al. Citation2016; Wang et al., Citation2016; Huang et al. Citation2018). The overall base composition is 31.13% A, 29.52% C, 24.46% T and 14.89% G. The average length of 13 PCGs genes is 875 bp. All the protein-coding genes use the initiation codon ATG except COI uses GTG, which is quite common in vertebrate mtDNA (Lin et al. Citation2018; Peng et al. Citation2019). All of the nodes were inferred with strong support by the NJ analysis. The phylogenetic tree showed that C. pectoralis clustered with C. brachyrhynchos and then together with other two crows in family Passeriformes (). In all, the mitochondrial genome reported here would be useful in the current understanding of the phylogeny and evolution of Passeriformes. We must pay more attention on Corvus species for lacking genetic information.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Haring E, Däubl B, Pinsker W, Kryukov A, Gamauf A. 2012. Genetic divergences and intraspecific variation in corvids of the genus Corvus (Aves: Passeriformes: Corvidae) – a first survey based on museum specimens. J Zool Syst Evol Res. 50:230–246.
- Huang T, Peng J, Zhao YL, Xu ZG. 2018. The complete mitochondrial genome of Pelecanus occidentalis (Pelecaniformes: Pelecanidae) and its phylogenetic analysis. Mitochondrial DNA Part B. 3:782–783.
- IUCN. 2019. The IUCN red list of threatened species [M]//V. 2019-2. http://www.iucnredlist.org.
- John E, Andrea KT, Irem S, Isao N. 2015. Patterns of evolution of MHC class II genes of crows (Corvus) suggest trans-species polymorphism. Peerj. 3:e853.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870.
- Lin Q, Jiang GT, Dai QZ, Li C. 2018. The complete mitochondrial genome of the Mayang white goose and its phylogenetic analyses. Mitochondrial DNA Part B. 54:1493–1497.
- Londei T. 2013. Alternation of clear‐cut colour patterns in Corvus crow evolution accords with learning‐dependent social selection against unusual‐looking conspecifics. IBIS. 155:632–634.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One. 7:e30619.
- Peng SM, Lin Q, Dai QZ, He X, Jiang GT. 2019. The complete mitochondrial genome of the Zhijiang duck and its phylogenetic analyses. Mitochondrial DNA Part B. 4:139–140.
- Ren T, Liang SR, Zhao AY, He K. 2016. Analysis of the complete mitochondrial genome of the Zhedong White goose and characterization of NUMTs: reveal domestication history of goose in China and Euro. Gene. 577:75–81.
- Wang JH, Liu G, Zhou LZ, Qing H, Li LY, Li B, Zhang LL. 2016. Complete mitochondrial genome of Tundra swan Cygnus columbianus jankowskii (Anseriformes: Anatidae). Mitochondrial DNA. 27:90–91.
- Zhang W, Zhao YL, Yang GY, Tang YC, Xu ZG. 2017. Characterization of the complete chloroplast genome sequence of Camellia oleifera in Hainan, China. Mitochondrial DNA Part B. 2:843–844.