Abstract
The long-legged fly Medetera sp. (Genbank accession number: MN604695) belongs to the subfamily Medeterinae of Dolichopodidae. The mitogenome of Medetera sp. was sequenced, the first representative of the mitogenome of the subfamily. The mitogenome is 14,740 bp totally, consisting of 13 protein-coding genes, two rRNAs, and 22 transfer RNAs. All genes have similar locations and strands with that of other published species of Dolichopodidae. The nucleotide composition biases toward A and T, which together made up 67.6% of the entirety. Bayesian inference analysis strongly supported the monophyly of Dolichopodidae. It suggested that subfamily Medeterinae is the sister group of subfamily Rhaphiinae.
Medeterinae belongs to the family Dolichopodidae, and the Medetera Fischer von Waldheim, 1819 is a larger genus of Medeterinae with 355 extant species and seven fossil species known from the world (Yang et al. Citation2006; Yang et al. Citation2011; Grichanov Citation2017).
The adult specimens of Medetera sp. (Genbank accession number: MN604695) used for this study were collected from Wang Lang Nature Reserve (32°91′52″N, 104°16′38″E, 2403 m) of Sichuan Province in China in 2016. The specimens of Medetera sp. were deposited in the Entomological Museum (Accession Number: D-M-1) of China Agricultural University (CAU). The total genomic DNA was extracted from the whole body (except head) of the specimen using the QIAamp DNA Blood Mini Kit (Qiagen, Germany) and stored at −20 °C until needed. The nearly complete mitogenome of Medetera sp. is 14,740 bp. It encoded 13 PCGs, 22 tRNA genes, two rRNA genes and the control region could not be sequenced entirely in this study and were similar with related reports before (Kang et al. Citation2016; Li et al. Citation2016; Wang, Ding, et al. Citation2016; Wang, Li, et al. Citation2016; Wang, Liu, et al. Citation2016; Wang, Wang, et al. Citation2016; Li et al. Citation2017; Zhou et al. Citation2017; Qilemoge, Gao, et al. Citation2018; Qilemoge, Zhang, et al. Citation2018; Hou et al. Citation2019; Qilemoge et al. Citation2019). The nucleotide composition of the mitogenome was biased toward A and T, with 67.6% of A + T content (A = 37.8%, T = 29.9%, C = 20.8%, and G = 11.6%). The A + T content of PCGs, tRNAs, and rRNAs is 65.3%, 72.8%, and 76.2%, respectively. The total length of all 13 PCGs of Medetera sp. is 11,213 bp. All PCGs in Medetera sp. utilize the conventional translational start codons for invertebrate mtDNA. For example, six PCGs (COII, COIII, ATP6, ND4, ND4L, and CYTB) initiated with ATG codons, two PCGs (ND3 and ND5) initiated ATT codons, three PCGs (ND2, ND1, and ND6) initiated ATA codons, ATP8 initiated ATC codon, and COI initiated TCG codon. Eight PCGs used the typical termination codons TAA and three PCGs (ND1, ND3, and CYTB) used TAG in Medetera sp.
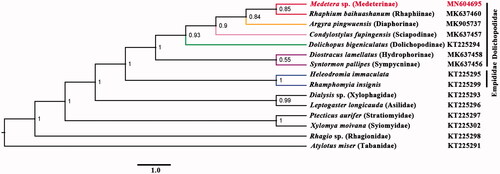
Phylogenetic analysis was performed based on the nucleotide sequences of 13 PCGs and two rRNAs from 15 Diptera species. Bayesian (BI) analysis () showed that monophyletic Empidoidea was assigned to be the sister group to the clade of Xylophagidae and Asilidae. For the phylogeny of Empidoidea, monophyletic Empididae was assigned to the sister to monophyletic Dolichopodidae. For the phylogeny of Dolichopodidae, Medeterinae was assigned to the sister of Rhaphiinae. The phylogenetic relationship within Empidoidea is very clear: Empididae + (Hydrophorinae + Sympycninae) + (Dolichopodinae + (Sciapodinae + (Diaphorinae + (Rhaphiinae + Medeterinae))))). The position of Medeterinae was also supported by the morphological study (Yang et al. Citation2011). The mitogenome of Medetera sp. could provide important information for the further studies of Dolichopodidae phylogeny.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Grichanov IY. 2017. Alphabetic list of generic and specific names of predatory flies of the epifamily Dolichopodidae (Diptera) 2nd ed. Plant Prot News, Suppl. 23:188–191.
- Hou P, Qilemoge Li X, Yang D. 2019. The mitochondrial genome of Syntormon pallipes (Diptera: Dolichopodidae). Mitochondrial DNA B. 4:1605–1606.
- Kang Z, Li X, Yang D. 2016. The complete mitochondrial genome of Dixella sp. (Diptera: Nematocera, Dixidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1528–1529.
- Li X, Ding S, Hou P, Liu X, Zhang C, Yang D. 2017. Mitochondrial genome analysis of Ectophasia rotundiventris (Diptera: Tachinidae). Mitochondrial DNA B. 2(2):457–458.
- Li X, Wang Y, Su S, Yang D. 2016. The complete mitochondrial genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1435–1436.
- Qilemoge Q, Gao S, Tang C, Wang N, Yang D. 2018. The mitochondrial genome of Diostracus lamellatus (Diptera: Dolichopodidae). Mitochondrial DNA B. 3(2):346–347.
- Qilemoge Q, Zhang J, Yang D. 2018. The mitochondrial genome of Rhaphium baihuashanum (Diptera: Dolichopodidae). Mitochondrial DNA B. 3(2):976–977.
- Qilemoge Q, Zhang C, He D, Zhang J, Yang D. 2019. The complete mitochondrial genome of Argyra pingwuensis (Diptera: Dolichopodidae). Mitochondrial DNA B. 4:2322–2323.
- Wang K, Ding S, Yang D. 2016. The complete mitochondrial genome of a stonefly species, Kamimuria chungnanshana Wu, 1948 (Plecoptera: Perlidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3810–3811.
- Wang K, Li X, Ding S, Wang N, Mao M, Wang M, Yang D. 2016. The complete mitochondrial genome of the Atylotus miser (Diptera: Tabanomorpha: Tabanidae), with mitochondrial genome phylogeny of lower Brachycera (Orthorrhapha). Gene. 586(1):184–196.
- Wang K, Wang Y, Yang D. 2016. The complete mitochondrial genome of a stonefly species, Togoperla sp. (Plecoptera: Perlidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(3):1703–1704.
- Wang Y, Liu X, Yang D. 2016. The complete mitochondrial genome of a fishfly, Dysmicohermes ingens (Chandler) (Megaloptera: Corydalidae: Chauliodinae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1092–1093.
- Yang D, Zhang L, Wang M, Zhu Y. 2011. Fauna Sinica Insecta. Vol. 53. Diptera Dolichopodidae. Beijing: Science Press.
- Yang D, Zhu Y, Wang M, Zhang L. 2006. World catalog of Dolichopodidae (Insecta: Diptera). Beijing: China Agricultural University Press.
- Zhou Q, Ding S, Li X, Zhang T, Yang D. 2017. Complete mitochondrial genome of Allognosta vagans (Diptera, Stratiomyidae). Mitochondrial DNA B. 2(2):461–462.