Abstract
In the present study, the complete mitogenome sequence of a Acanthorhodeus chankaensis Dybowsky from Cao’e River was sequenced and identified. The assembled mitogenome of A. chankaensis is 16,676 bp in length, it contains 22 transfer-RNA genes, 13 protein-coding genes, 2 ribosomal-RNA genes, and 2 non-coding regions. It shows conserved gene arrangement with other Cyprinidae fishes. The overall nucleotide composition of A. chankaensis mitogenome sequence is A: 28.96%, G: 17.11%, T: 27.46%, and C: 26.47%. The phylogenetic analysis showed that the complete mitogenome could contribute to the phylogenetic analyses and population genetics study of A. chankaensis and Acanthorhodeus fish.
Acanthorhodeus chankaensis Dybowsky, a small-sized fish distributed widely in various water systems in China (Chen et al. Citation2005; Chen, Zhao, et al. Citation2013; Meng et al. Citation2016; Wang et al. Citation2019). It is part of the food web in natural waters because of its large population and plays an important role in maintaining water ecosystem balance. Except for its edible value, it also has ornamental value due to their marital color during the breeding season (Zhao et al. Citation2010; Wang et al. Citation2015, Wang Citation2019). However, A. chankaensis is suffering from rapid population reductions because of pesticide application, water pollution and increasing capture pressure (Wang et al. Citation2015). Cao’e River is the main channel in East China (Xie and Pan Citation2013; Han, Citation2018). Although A. chankaensis is a common fish in China, knowledge about it in Cao’e River is largely unknown.
The mitochondrial genome (mitogenome) sequence has compact gene arrangement, short coalescence time, and rapid evolutionary rate (Habib et al. Citation2012; Yang et al. Citation2018), which could provide useful data for phylogenetic analyses, population genetics and evolution study (Chen, Zhao, et al. Citation2013; Wang et al. Citation2015; Yang et al. Citation2015; Yu et al. Citation2019).
In the present study, a new mitochondrial genome of A. chankaensis (GenBank accession no. MN683735) was sequenced and annotated. Acanthorhodeus chankaensis was collected from Cao’e River, Zhejiang Province in China (30°00′08.4′′N, 120°52′49.7′′E) and kept in 99% ethanol in Shaoxing Aquatic Service Platform (SXAF191125). The total genomic DNA of A. chankaensis was isolated and was quantified by ultra-micro spectrophotometry. The PCR amplification was carried out using the following protocols: initial denaturation at 93 °C for 4 min, followed by 38 cycles (94 °C for 45 s, 50–52 °C for 35 s, and 72 °C for 80 s) and 1 final cycle of 6 min at 72 °C.
The complete mitochondrial genome of A. chankaensis is 16,676 bp in length, it contains a conserved arrangement with other Cyprinidae fishes (Wei et al. Citation2015; Yang et al. Citation2015), which include 22 transfer RNA genes, 13 protein-coding genes (PCDs), 2 ribosomal RNA genes, and 2 non-coding regions (control region and D-loop).
The nucleotide composition of A. chankaensis mitogenome sequence is A: 28.96%, G: 17.11%, T: 27.46%, and C: 26.47%. A similar A + T bias (56.42%) was seen with other vertebrate mitogenomes (Liu and Cui Citation2009; Wan et al. Citation2013; Pan et al. Citation2015).
The total sequence length of the PCD genes is 11,408 bp. The total length of the tRNA genes is 1562 bp, their sizes ranged from 68 bp (tRNACys) to 76 bp (tRNALys and tRNALeu). The 12S and 16S rRNA genes are 958 bp and 1679 bp in length, respectively. Similar to other vertebrate mitogenomes, these two genes are located between the genes tRNAPhe and tRNALeu(UUR) and are separated by the gene for tRNAVal.
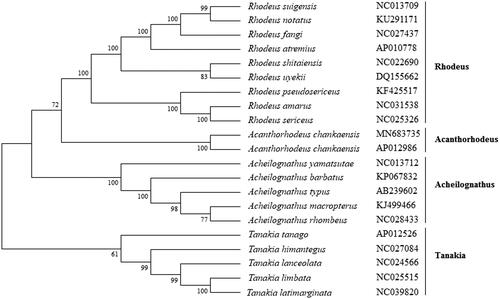
The phylogenetic analysis showed that A. chankaensis was firstly clustered with another reported A. chankaensis (Lv et al. Citation2015), and then was clustered in the genus Acanthorhodeus with other Acanthorhodeus fishes (). While it showed distant kinship with other Rhodeus fishes. This study provides useful data to phylogenetic analyses and population genetics of Acanthorhodeus fishes.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Chen ST, Zhao RY, Qi DF, Fan XL, Lei HZ, Lin ZH. 2013. Sexual dimorphism in morphological traits and female individual fecundity of Acanthorhodeus chankaensis Dybowsky. J Shanghai Jiaotong Univ (Agric Sci). 31(5):61–66.
- Chen XH, Ni Y, Wu HL. 2005. Fish of genus Rhodeus agassiz in Jiangsu province, China. Mar Fisheries. 27(2):89–97.
- Habib M, Lakra WS, Mohindra V, Lal KK, Punia P, Singh RK, Khan AA. 2012. Assessment of ATPase 8 and ATPase 6 mtDNA sequences in genetic diversity studies of Channa marulius (Channidae: Perciformes). Proc Natl Acad Sci, India, Sect B Biol Sci. 82(4):497–501.
- Han FF. 2018. Analysis on recent river bed change of Zhakou-Caoejiang estuary in Qiantang Jiang River. Shanghai Water. 34(1):1–5.
- Liu Y, Cui Z. 2009. The complete mitochondrial genome sequence of the cutlass fish Trichiurus japonicus (Perciformes: Trichiuridae): Genome characterization and phylogenetic considerations. Mar Genom. 2(2):133–142.
- Lv WH, Cao DC, Wu J, Zheng XH, Sun XW. 2015. The complete mitochondrial genome sequence of Khanka spiny bitterling (Acanthorhodeus chankaensis). Mitochondrial DNA. 26(6):842–843.
- Meng XL, Zhou CJ, Li XJ, Kong XH, Qiu ZP, Zhang MC, Xiong ZY, Nie GX. 2016. Investigation on fishery resources in Luoshan county of Xinyang. Hannan Fisheries. 2:22–25.
- Pan D, Wei M, Wan Q, Tao G, Ma Z, Wang H. 2015. The complete mitochondrial genome of Pseudobagrus trilineatus (Siluriformes: Bagridae). Mitochondrial DNA. 26(6):893–894.
- Wan Q, Tao G, Cheng Q, Chen Y, Qiao H. 2013. The complete mitochondrial genome sequence of Pseudobagrus ussuriensis (Siluriformes: Bagridae). Mitochondrial DNA. 24(4):333–335.
- Wang JG, Wang Q, Du HY, Xu XB, Chen XJ, Xiong LW, Zhu YG, Feng Q. 2015. Study on biological characteristics of Acanthorhodeus chankaensis in Lixia River basin. Jiangsu Agri Sci. 43(9):256–259.
- Wang JL, Chen QH, Lu WZ, Yuan SQ, Hu QX, Cen SS, Zhou CJ, Meng XL, Nie GX, Gu QH. 2019. Morphological differences among five species in Rhodeinae in Huaihe River basin in Henan province. Acta Hydrobiologica Sinica. 43(1):123–132.
- Wang YC. 2019. Investigation and analysis of fish resources at the confluence of the Weihe River and the Yellow River. Acta Ecologiae Animalis Domastici. 40(1):41–45.
- Wei M, Pan D, Yang Y, Tao G, Ding S, Wan Q. 2015. The complete mitochondrial genome of Chinese lizard gudgeon, Saurogobio dabryi (Cypriniformes: Cyprinidae). Mitochondrial DNA. 26(6):945–946.
- Xie DF, Pan CH. 2013. A preliminary study of the turbulence features of the tidal bore in the Qiantang River, China. J Hydrodyn. 25(6):903–911.
- Yang HR, Zhang JE, Xia J, Yang JZ, Guo J, Deng ZX, Luo MZ. 2018. Comparative characterization of the complete mitochondrial genomes of the three apple snails (Gastropoda: Ampullariidae) and the phylogenetic analyses. IJMS. 19(11):3646.
- Yang SB, Mi ZX, Tao G, Liu XF, Wei M, Wang H. 2015. The complete mitochondrial genome sequence of Margaritiana dahurica Middendorff. Mitochondrial DNA. 26(5):716–717.
- Yu P, Yang X, Zhou W, Yang W, Zhou L, Liu X, Wan Q, Zhang J. 2019. Comparative mitogenomic and phylogenetic analysis of Apalone spinifera and Apalone ferox (Testudines: Trionychidae). Genetica. 147(2):165–176.
- Zhao ZY, Jiang YZ, Fang XZ, Zhou Y. 2010. Biological characteristics and ornamental value of Rhodeus fish. Bull Biol. 45(4):7–9.