Abstract
Northern snakehead, Channa argus, is a commercially important food fish species in China. In the present study, the complete mitochondrial genome of C.argus from the Baima Hu Lake was characterized. It is 16,558 bp in length, consist of 22 tRNA genes, 13 PCD genes, 2 rRNA genes, and 1 D-loop region. The overall base composition of the C. argus mitogenome is 27.26% A, 24.21% T, 31.58% C and 16.95% G, exhibits a similar AT bias (51.47%) feature to other vertebrate mitogenomes. The phylogenetic analysis showed that C. argus clustered in genus Channa. The present resultes provide useful information to population genetics and conservation biology studies of Channa fishes.
Northern snakehead Channa argus, a benthic carnivorous fish (Yue et al. Citation1996; Ermolenko and Besprozvannykh Citation2008; Dong et al. Citation2014a; Duan et al. Citation2018), which is distributed widely in various water systems of China, North Korea, Japan, Southeast Asia, India and Russian (Odenkirk and Owens Citation2005; Hossain et al. Citation2008; Nguyen et al. Citation2012; Dong et al. Citation2014b; Densmore et al. Citation2016). It is a commercially important fish species in China known for its fast growth, high meat content with few bone spurs, tolerance to water pollution and diseases (Ishimatsu and Itazawa Citation1983; Sagada et al. Citation2017; Zhou et al. Citation2018; Chen et al. Citation2019; Li et al. Citation2019; Fang et al. Citation2019).
The Baima Hu Lake is a small lake in Shangyu district, Shaoxing city, East China. Although C.margus is artificially cultured for a long time in China (Wang et al. Citation2012, Citation2019), little is known about it in the Baima Hu Lake.
The mitogenome could provide useful data for population genetics and conservation biology studies of vertebrate fish, due to rich signals from its sequence and conserved gene arrangement (Liu and Cui Citation2009; He et al. Citation2014; Li et al. Citation2016; Zhu et al. Citation2019).
Herein, the complete mitogenome of C. argus (GenBank accession no. MN781664) was characterized. Channa margus individual was sampled from the Baima Hu Lake, Zhejiang Province of China (33°13′47.7″N 119°08′49.4″E), and was kept in 99% ethanol in the Aquatic Service Platform of Shaoxing (accession no. SXAF20200508). The PCR fragments were amplified and sequcenced.
The complete mitochondrial genome of C. argus is 16,558 bp in length, consists of 22 tRNA genes, 13 protein-coding genes (PCDs), 2 rRNA genes, and 2 non-coding regions. The total length of the protein-coding gene sequences is 11,900 bp. Except for the ND6 being encoded on the L-strand, all the other PCD genes (ND1-5, ND4L, COXI-III, ATP6, ATP8, and CytB) are encoded on the H-strand. The total length of all tRNA genes is 1540 bp, varying from 65 bp (tRNACys) to 75 bp (tRNALys). The 12S rRNA gene (946 bp) and 16S rRNA (1687 bp) gene are located between two tRNA genes (tRNAPhe and tRNALeu(UUR), and are separated by tRNAVal gene. The length of D-loop region is 992 bp. The gene structure and arrangement of C. argus are very similar to other vertebrate species (Chen et al. Citation2013; Yang et al. Citation2015).
The overall base composition of the C. argus mitogenome is 27.26% A, 24.21% T, 31.58% C and 16.95% G, respectively, which exhibits a similar AT bias (51.47%) feature to other vertebrate mitogenomes (Yang et al. Citation2015; Cui et al. Citation2019).
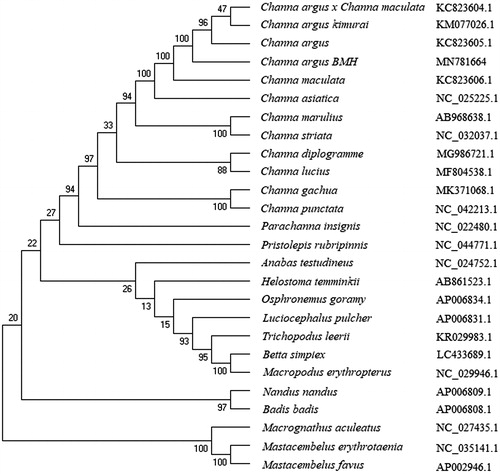
The phylogenetic analysis showed that C. argus is clustered with other C. argus reported (Wang and Yang Citation2011), and clustered together with other Channa fishes, including Channa asiatica, Channa diplogramme, Channa gachua, Channa lucius, Channa maculata, Channa marulius, Channa striata, and Channa punctata (). But, it showed distant kinship with other Cyprinidae fishes. This study provides useful data to population genetics and conservation biology studies of Channa fishes.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available at NCBI (https://www.ncbi.nlm.nih.gov), GenBank accession no. MN781664. And the data that support the findings of this study are also available from the corresponding author, Dr. Yang, upon reasonable request.
Additional information
Funding
References
- Chen C, Yang B, Abbas Raza SH, Zhang D, Wu T, Zhang Z, Ullah I, Khan R, Yang G, Wang C, et al. 2019. Role of Myeloperoxidase of northern snakehead (Channa argus) in Aeromonas veronii infection. Microb Pathog. 135:103622.
- Chen X, Zhou ZM, Chen ZJ, Ai WM. 2013. Complete mitochondrial genome of Sarcocheilichthys parvus (Cypriniformes, Cyprinidae). Mitochondrial DNA. 24(2):97–98.
- Cui LL, Gao HT, Miao XJ, Li ML, Li GH, Xu GF, Wu JJ, Hu W, Lu SX. 2019. The complete mitochondrial genome of Pareuchiloglanis myzostoma (Teleostei, Siluriformes). Mitochondrial DNA Part B. 4(2):3626–3627.
- Densmore CL, Iwanowicz LR, Henderson AP, Iwanowicz DD, Odenkirk JS. 2016. Mycobacterial infection in Northern snakehead (Channa argus) from the Potomac River catchment. J Fish Dis. 39(6):771–775.
- Dong XP, Mu SM, Zhou N, Kang XJ, Bai JJ. 2014a. Gentic diversity of mitochondrial DNA D-Loop sequence of Channa argu in Baiyangdian. J Hebei Univ (Nat Sci Edn). 34(2):201–206.
- Dong XP, Mu SM, Zhou N, Kang XJ, Luo Q, Bai JJ. 2014b. Structure analysis of mt DNA D-Loop region and the genetic diversity of Channa argus in different populations. J Fish China. 38(9):1277–1285.
- Duan T, Shi CC, Zhou J, Lv X, Li YL, Luo YP. 2018. How does the snakehead Channa argus survive in air? The combined roles of the suprabranchial chamber and physiological regulations during aerial respiration. Biology Open 8. 7(2):bio029223.
- Ermolenko AV, Besprozvannykh VV. 2008. Parasite fauna of the snakehead Channa argus warpachowskii from Primorsky Krai. Parazitologia. 42(4):325–329.
- Fang HH, Xie JJ, Liao SY, Guo TY, Xie SW, Liu YJ, Tian LX, Niu J. 2019. Effects of dietary inclusion of shrimp paste on growth performance, digestive enzymes activities, antioxidant and immunological status and intestinal morphology of hybrid snakehead (Channa maculata ♀ × Channa argus ♂). Front Physiol. 10:1027.
- He LP, Yang SB, Zheng DH, Li C, Tao G, Wei M, Wang H. 2014. Complete mitochondrial genome of Pseudorasbora elongata (Cypriniformes: Cyprinidae): sequencing and analysis. Mitochondrial DNA. 25(6):433–434.
- Hossain M, Latifa G, Rahman M. 2008. Observations on induced breeding of snakehead murrel, Channa striatus (Bloch, 1793). Int J Sustainable Crop Prod. 3:65–68.
- Ishimatsu A, Itazawa Y. 1983. Blood oxygen levels and acid-base status following air exposure in an air-breathing fish, Channa argus: the role of air ventilation. Comparative Biochem Physiol Part A. 74(4):787–793.
- Li C, He LP, Chen C, Cai LC, Chen PP, Yang SB. 2016. Sequencing, description and phylogenetic analysis of the mitochondrial genome of Sarcocheilichthys sinensis sinensis (Cypriniformes: Cyprinidae). Mitochondrial DNA A. 27(2):1056–1057.
- Li MY, Guo WQ, Guo GL, Zhu XM, Niu XT, Shan XF, Tian JX, Wang GQ, Zhang DM. 2019. Effect of sub-chronic exposure to selenium and Allium mongolicum Regel flavonoids on Channa argus: bioaccumulation, oxidative stress, immune responses and immune-related signaling molecules. Fish Shellfish Immunol. 91:122–129.
- Liu Y, Cui Z. 2009. The complete mitochondrial genome sequence of the cutlass fish Trichiurus japonicus (Perciformes: Trichiuridae): genome characterization and phylogenetic considerations. Mar Geonom. 2(2):133–142.
- Nguyen TC, Li YC, Makouloutou P, Jimenez LA, Sato H. 2012. Posthodiplostomum sp. Metacercariae in the trunk muscle of northern snakeheads (Channa argus) from the Fushinogawa River, Yamaguchi, Japan. J Vet Med Sci. 74(10):1367–1372.
- Odenkirk J, Owens S. 2005. Northern snakeheads in the tidal Potomac River system. Trans Am Fish Soc . 134(6):1605–1609.
- Sagada G, Chen JM, Shen BQ, Huang AX, Sun LH, Jiang JH, Jin CH. 2017. Optimizing protein and lipid levels in practical diet for juvenile northern snakehead fish (Channa argus). Anim Nutrition. 3(2):156–163.
- Wang J, Yang G. 2011. The complete mitogenome of the snakehead Channa argus (Perciformes: Channoidei): genome characterization and phylogenetic implications. Mitochondrial DNA. 22(4):120–129.
- Wang Q, Wang W, Huang Q, Zhang Y, Luo Y. 2012. Effect of meal size on the specific dynamic action of the juvenile snakehead (Channa argus). Comparative Biochem Physiol Part A. 161(4):401–405.
- Wang L, Xie N, Shen Y, Ye B, Yue GH, Feng X. 2019. Constructing high-density genetic maps and developing sexing markers in northern snakehead (Channa argus). Mar Biotechnol. 21(3):348–358.
- Yang S, Mi Z, Tao G, Liu X, Wei M, Wang H. 2015. The complete mitochondrial genome sequence of Margaritiana dahurica Middendorff. Mitochondrial DNA. 26(5):716–717.
- Yue CX, Gao ST, Deng FJ, Xu HM, Liu ZL. 1996. A Histological study on the ovary development of channa argus. J Wuhan Univ (Nat Sci Edn). 42(2):225–232.
- Zhou AG, Xie SL, Wang ZL, Chen YF, Zhang Y, Fan LF, Zeng F, Zou JX. 2018. HSP60 expression profile under different extreme temperature stress in albino northern snakehead, Channa argus. Cell Stress Chaperones. 23(4):791–796.
- Zhu K, Gao Y, Yuan P, Cao P, Ying X, Tao H, Liu B. 2019. The complete mitochondrial genome of Ostorhinchus fleurieu (kurtiformes: Apogonidae) and phylogenetic studies of apogoninae. Mitochondrial DNA Part B. 4(2):3691–3692.