Abstract
The mitogenome of Ilythea japonica was sequenced. The mitogenome was 16,256 bp totally, consisting of 13 protein-coding genes, two rRNAs, and 22 transfer RNAs. The nucleotide composition biases toward A and T is 77.1% of the entirety. All PCGs start with ATN codons except COI and ND1, and end with TAA or incomplete stop codon. Phylogenetic analyses based on 10 dipteran species supported the monophyly of Ephydroidea and the relationship of Opomyzoidea + (Ephydroidea + (Lauxanioidea + (Sphaeroceroidea + (Sciomyzoidea + +Tephritoidea))))
Introduction
Ephydridae is a family with body size from minute to moderate (1.0–11.0 mm). Adults of this family are often dull and dark colored, but unusually diverse in body structure, vestiture, and ornamentation (Mathis and Zatwarnicki Citation1998). The ephydrid flies, which have about 2000 described species worldwide, are placed in superfamily Ephydroidea (Pape et al. Citation2011). Species of Ilythea, which feed on diatoms, are common in stream and wetland habitats (Foote Citation1995).
The adult specimens of Ilythea japonica (accession number: YDEPHY-ILY) used for this study were collected from Houzhenzi (33.8520°N, 107.8429°E, 1278 m), Zhouzhi, Shaanxi, China. The specimens were identified by Liang Wang and deposited in the Entomological Museum of China Agricultural University (CAU).
The genomic DNA was extracted from adult’s whole body using the DNeasy DNA Extraction kit (TIANGEN) and stored at −20 °C refrigerator. The library was sequenced on an Illumina HiSeq 2500. The position of all tRNA genes was confirmed using tRNAscanSE 2.0 (Lowe and Chan Citation2016). The nearly complete mitogenome of I. japonica (MT527723) was 16,256 bp in length and consisted of 13 typical invertebrate PCGs, 22 transfer RNA genes, two rRNA genes (12S and 16S), and part control region, which were similar to other flies reported before (Li et al. Citation2016; Zhou et al. Citation2017; Qilemoge et al. Citation2018; Ren et al. Citation2019). The nucleotide composition of the mitogenome was biased toward A and T, with 77.1% of A + T content (A = 39.8%, T = 37.3%, C = 14.4%, G = 8.5%). Among the protein-coding genes, six genes took the start codon of ATG, five genes used ATT as start codon, while COI gene and ND1 gene got ACG and TTG, respectively. The termination codon of these protein-coding genes had three types (nine genes used TAA, CYTB gene used TAG, three genes used T + tRNA).
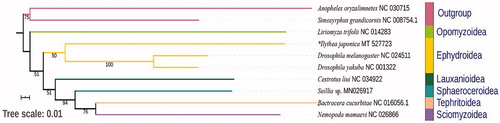
There are data of nine species retrieved from NCBI and one new sequenced datum used in phylogenetic analysis. The genbank accession numbers are listed as follows: Anopheles oryzalimnetes NC_030715, Bactrocera cucurbitae NC_016056.1, Ceratitis capitata NC_000857, Drosophila melanogaster NC_024511, Drosophila yakuba NC_001322, *Ilythea japonica MT527723, Liriomyza trifolii NC_014283, Nemopoda mamaevi NC_026866, Simosyrphus grandicornis NC_008754.1, Suillia sp. MN 026917. Thirteen protein-coding genes (PCGs) were used to reconstruct phylogenetic relationship with maximum-likelihood (ML) method. The topology was given and bootstrap support numbers are shown in . ML analysis revealed that Ephydroidea was monophyletic. The higher-level relationship of Opomyzoidea + (Ephydroidea + (Lauxanioidea + (Sphaeroceroidea + (Sciomyzoidea + Tephritoidea)))) was supported.
Figure 1. The phylogenetic tree of ML analysis based on 13PCGs; “*” indicated new sequenced datum in this study.
The complete mitochondrial genome of I. japonica provides the valuable information for future genetic and evolutionary studies of family Ephydridae and superfamily Ephydroidea.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT527723.
Additional information
Funding
References
- Foote BA. 1995. Biology of shore flies. Annu Rev Entomol. 40(1):417–442.
- Li X, Wang Y, Su S, Yang D. 2016. The complete mitochondrial genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA A. 27(2):1435–1436.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. (Web Server issue)
- Mathis W, Zatwarnicki T. 1998. Family 3.49 Ephydridae. In: Papp L, Darvas B, editors. Contributions to a manual of Palaearctic Diptera. Vol. 3. Budapest: Science Herald; p. 537–570.
- Pape T, Blagoderov V, Mostovski MB. 2011. Order Diptera Linnaeus, 1758. Zootaxa. 3148(1):222–229.
- Qilemoge , Gao S, Tang C, Wang N, Yang D. 2018. The mitochondrial genome of Diostracus lamellatus (Diptera: Dolichopodidae). Mitochondrial DNA B. 3(2):346–347.
- Ren J, Yang Q, Gao S, Pan Z, Chang W, Yang D. 2019. The mitochondrial genome of Limonia phragmitidis (Diptera Limoniidae). Mitochondrial DNA B. 4(1):719–720.
- Zhou Q, Ding S, Li X, Zhang T, Yang D. 2017. Complete mitochondrial genome of Allognosta vagans (Diptera, Stratiomyidae). Mitochondrial DNA B. 2(2):461–462.

