Abstract
The complete mitochondrial genome of Ancherythroculter nigrocauda was determined in this study. It contained 1 replication origin, 1 control region (D-Loop), 2 rRNA genes, 13 PCGs, and 22 tRNA genes with the base composition 31.40% A, 25.00% T, 27.60% C, and 16.00% G. Here, we compared this newly determined mitogenome with another one from the same species reported before. The variable sites and the genetic distances between the two mitogenomes were 608 bp and 0.038, respectively. The results from the phylogenetic analysis showed that the genus Ancherythroculter is not a monophyletic group and Ancherythroculter nigrocauda demonstrates a close relationship with Chanodichthys dabryi.
Ancherythroculter nigrocauda (Cypriniformes, Cyprinidae, Oxygastrinae) is an endemic species in China, distributed in Sichuan and Chongqing (Froese and Pauly Citation2019). It has high economic value and nutritional value. Here, the complete mitochondrial genome sequence of A. nigrocauda was determined (GenBank accession No. MT588183) and was compared with another A. nigrocauda mitogenome data reported before (Wan et al. Citation2013). The specimens (voucher no. SWU20110226005) were collected from Jialing River (29.97 N, 106.28 E), Hechuan, Chongqing, China, and were stored in the museum of the Key Laboratory of Freshwater Fish Reproduction and Development (Ministry of Education, Southwest University, Chongqing, China). We extracted DNA and designed primers according to the method of Wang et al. (Citation2011). The mitochondrial genome is annotated via the MITOS WebServer (Bernt et al. Citation2013).
The complete mitochondrial genome of A. nigrocauda was a circular molecule with 16,620 bp in length. It contained 1 replication origin, 1 control region (D-Loop), 2 rRNA genes, 13 PCGs, and 22 tRNA genes. Most of the genes were encoded on H-strand, while ND6 and 8 tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu, and tRNA-Pro) were encoded on L-strand. The overall nucleotide composition was 31.40% A, 25.00% T, 27.60% C, and 16.00% G, with a slight AT bias. All the mitochondrial PCGs in the A. nigrocauda use the standard ATG start codon, except for CO1, which utilizes GTG. Six PCGs (ND1, COI, ATP6, CO III, ND4L, and ND5) contain TAA stop codon, five PCGs (ND2, ATP8, ND3, ND4, and ND6) contain TAG stop codon and two PCGs (COII, Cytb) contain the incomplete stop codon T–. The arrangement of complete mitochondrial genome of A. nigrocauda is basically the same as the previous research (Li et al. Citation2012; Wan et al. Citation2013).
Comparing with another mitogenome of A. nigrocauda, the length of it was 16,623 bp (accession no. KC513573; Wan et al. Citation2013). The genetic distance between the two mitogenomes was 0.038. Two indels and 608 variable sites exist in between the two mitogenomes. Among these variable sites, 566 transitions (93%) and 42 transversions (7%) were found.
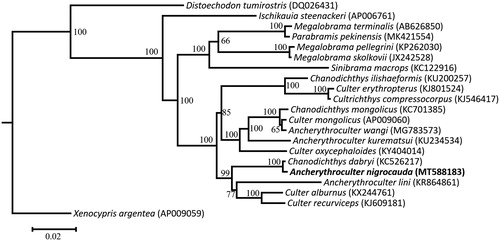
To confirm the phylogenetic position of A. nigrocauda among the genus Ancherythroculter, phylogenetic analysis based on the complete mitogenome using maximum-likelihood was conducted using IQ-tree (Nguyen et al. Citation2015; Hoang et al. Citation2018). The ModelFinder was used to calculate the optimal nucleotide substitution model (Kalyaanamoorthy et al. Citation2017). Phylogenetic maximum likelihood tree is given (). The results from the analyses show that the genus Ancherythroculter is not a monophyletic group and A. nigrocauda demonstrates a close relationship with Chanodichthys dabryi.
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT588183.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Froese R, Pauly D, editors. 2019. FishBase version (08/2019). www.fishbase.org.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Li P, Xu D, Peng Z, Zhang Y. 2012. The complete mitochondrial genome of the spotted steed, Hemibarbus maculatus (Teleostei, Cypriniformes). Mitochondrial DNA. 23(1):34–36.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Wan Q, Chen Y, Cheng Q, Qiao H. 2013. The complete mitochondrial genome sequence of Ancherythroculter nigrocauda (Cypriniformes: Cyprinidae). Mitochondrial DNA. 24(6):627–629.
- Wang J, Li P, Zhang Y, Peng Z. 2011. The complete mitochondrial genome of Chinese rare minnow, Gobiocypris rarus (Teleostei: Cypriniformes). Mitochondrial DNA. 22(5–6):178–180.