Abstract
In this study, we announce the complete mitochondrial genome (mitogenome) of the Korean endemic firefly, Luciola unmunsana Doi, Citation1931. The full-length circular genome was 15,858 bp, with 77.94% A/T content. It contained the typical set of 37 metazoan genes: 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and 2 ribosomal RNA (rRNA) genes, as well as an A + T-rich region. The gene arrangement of the species is identical to that of the ancestral arrangement found in the majority of insects. The maximum-likelihood tree, built using all PCGs and two rRNAs via randomized accelerated maximum likelihood (RAxML) showed that L. unmunsana was grouped as a sister to L. curtithorax with the highest nodal support. However, another Luciola species clustered with the Aquatica species such that the genus Luciola was a non-monophyletic group. Therefore, more sampling is required to clarify the phylogeny of Luciola.
The genus Luciola Laporte, 1833 is a firefly (subfamily Luciolinae) (Stanger-Hall et al. Citation2007), and three species are found in Korea: L. lateralis, L. unmunsana, and L. papariensis (Kang Citation2012). Among them, L. unmunsana Doi Citation1931 (Coleoptera: Lampyridae), is an endemic species in Korea (Doi Citation1931). Luciola unmunsana and L. papariensis are nearly identical morphologically. A single morphological character, which can distinguish the two species, is the color pattern of the pronotum (Kang Citation2012). Recently, Han et al. (Citation2020) studied the phylogeny of the Luciola species to elucidate the species status of the two Korean Luciola species using the DNA barcode region. However, their results showed that the lineages of the two species split into several groups, although intergroup relationship was not distinct enough to consider each an independent species. In this study, we sequenced the complete mitochondrial genome (mitogenome) of L. unmunsana (GenBank accession number: MT134039) for subsequent mitogenome-based phylogenetic analysis.
An L. unmunsana adult was collected from Mt. Unmunsan, Cheongdo, Gyeongsangbuk-do Province in Korea (35°39′17.7″N, 128°57′57.4″E) and total DNA was extracted from two hind legs. The leftover DNA and the specimen were deposited at the Chonnam National University, Gwangju, Korea, under the accession number CNU12790. Full-length mitogenome sequence data for L. unmunsana were obtained via next-generation sequencing using the MGISEQ-2000 sequencing platform (MGI Tech Co. Ltd, Shenzhen, China). Genome construction was performed using de novo assembly. Owing to the precise nature of final genome sequence, no additional Sanger-based sequencing was conducted. Phylogenetic analysis was performed within the scope of Luciolinae with 15 available mitogenomes, including L. unmunsana, using randomized axelerated maximum likelihood (RAxML) (Stamatakis Citation2014). For the analysis, 13 protein-coding genes (PCGs) and 2 ribosomal RNA (rRNA) genes were aligned and a total length of 12,517 bp (excluding gaps) was analyzed using the substitution model, GTR + Gamma + I.
We assembled the 15,858-bp long complete mitogenome of L. unmunsana from 137,521,575 high-quality clean reads. The genome contained 13 PCGs, 22 transfer RNAs (tRNAs), 2 rRNAs, and 1 major non-coding A + T-rich region that was 1247 bp long. The overall A/T nucleotide composition of the L. unmunsana mitogenome was as follows: 75.82% in the 13 PCGs, 77.94% in the whole genome, 79.57% in tRNAs, 80.72% in srRNA, 82.68% in lrRNA, and 88.13% in the A + T-rich region. The gene arrangement of L. unmunsana was identical to that of the ancestral arrangement found in the majority of insects (Boore Citation1999). The L. unmunsana mitogenome had the shortest length (16,385 bp on average) among mitogenomes of 16 Luciolinae members, which ranged from 15,967 bp (Asymmetricata circumdata; Luan and Fu Citation2016) to 16,882 bp (L. curtithorax; Hu and Fu Citation2018a).
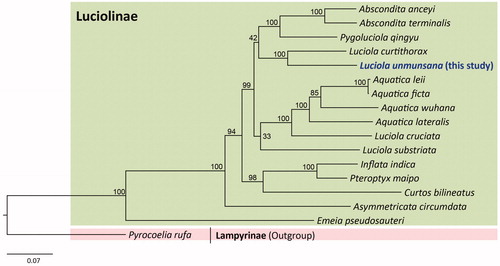
Luciola species comprises non-monophyletic groups, placing the current L. unmunsana as a sister to L. curtithorax with the highest nodal support, whereas other species of Luciola were grouped together with species of Aquatica (). Following Ballantyne and Lambkin’s (Citation2009) study, wherein the genera are presented as polyphyletic groups using 343 morphological characters, a new genus, Aquatica, was proposed for some species of Luciola using behavioral and morphological evidence (Fu et al. Citation2010). Nevertheless, additional phylogenetic revision with the inclusion of extended taxa is required to further clarify the phylogeny of Luciola.
Figure 1. Phylogenetic tree for the subfamily Luciolinae. The maximum likelihood (ML) method was applied using randomized accelerated maximum likelihood (RAxML) ver. 8.0.24 (Stamatakis Citation2014), which was incorporated into the cyberinfrastructure for phylogenetic research (CIPRES) Portal ver. 3.1 (Miller et al. Citation2010). A six optimal partitioning scheme and substitution model (GTR + Gamma + I) were determined using PartitionFinder 2 with the Greedy algorithm (Lanfear et al. Citation2012, Citation2014, Citation2016). Phylogenetic trees were visualized using FigTree ver. 1.42 (http://tree.bio.ed.ac.uk/software/figtree/). The numbers at each node represent bootstrap percentages of 1,000 pseudoreplicates by ML analysis. The scale bar indicates the number of substitutions per site. Lampyrinae (Pyrocoelia rufa, MH352481; Bae et al. Citation2004) is used as an outgroup. GenBank accession numbers are as follows: Abscondita anceyi, MH020192 (Hu and Fu Citation2018b); Abscondita terminalis, MK292092 (Chen et al. Citation2019); Pygoluciola qingyu, MK292093 (Chen et al. Citation2019); Aquatica leii, KF667531 (Jiao et al. Citation2015); Aquatica ficta, KX758085 (Wang et al. Citation2017); Aquatica wuhana, KX758086 (Wang et al. Citation2017); Luciola cruciata, AB849456 (Maeda et al. Citation2017a); Aquatica lateralis, LC306678 (Maeda et al. Citation2017b); Luciola curtithorax, MG770613 (Hu and Fu Citation2018a); Luciola substriata, KP313820 (Mu et al. Citation2016); Inflata indica, MH427718 (Sriboonlert and Wonnapinij Citation2019); Pteroptyx maipo, MF686051 (Fan and Fu Citation2017); Curtos bilineatus, MK292114 (Chen et al. Citation2019); Asymmetricata circumdata, KX229747 (Luan and Fu Citation2016); and Emeia pseudosauteri, MK292112 (Chen et al. Citation2019).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in Mendeley Data at http://dx.doi.org/10.17632/rh3cd4ztx7.1
Additional information
Funding
References
- Bae JS, Kim I, Sohn HD, Jin BR. 2004. The mitochondrial genome of the firefly, Pyrocoelia rufa: complete DNA sequence, genome organization, and phylogenetic analysis with other insects. Mol Phylogenet Evol. 32(3):978–985.
- Ballantyne LA, Lambkin C. 2009. Systematics of Indo-Pacific fireflies with a redefinition of Australasian Atyphella Olliff, Madagascan Photuroluciola Pic, and description of seven new genera from the Luciolinae (Coleoptera: Lampyridae). Zootaxa. 1997(1):1–188.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Chen X, Dong Z, Liu G, He J, Zhao R, Wang W, Peng Y, Li X. 2019. Phylogenetic analysis provides insights into the evolution of Asian fireflies and adult bioluminescence. Mol Phylogenet Evol. 140:106600.
- Doi H. 1931. A new species of Luciola from Korea, Luciola unmunsana, sp. nov. J Chosen Nat Hist Soc. 12:54–55.
- Fan Y, Fu X. 2017. The complete mitochondrial genome of the firefly, Pteroptyx maipo (Coleoptera: Lampyridae). Mitochondrial DNA Part B. 2(2):795–796.
- Fu XH, Ballantyne L, Lambkin CL. 2010. Aquatica gen. nov from mainland China with a description of Aquatica Wuhana sp nov (Coleoptera: Lampyridae: Luciolinae). Zootaxa. 2530(1):1–18.
- Han T, Kim SH, Yoon HJ, Park IG, Park H. 2020. Evolutionary history of species of the firefly subgenus Hotaria (Coleoptera, Lampyridae, Luciolinae, Luciola) inferred from DNA barcoding data. Contrib Zool. 89(2):127–119.
- Hu J, Fu X. 2018a. The complete mitochondrial genome of the firefly, Luciola curtithorax (Coleoptera: Lampyridae). Mitochondrial DNA Part B. 3(1):378–379.
- Hu J, Fu X. 2018b. The complete mitochondrial genome of the firefly, Abscondita anceyi (Olivier) (Coleoptera: Lampyridae). Mitochondrial DNA Part B. 3(1):442–443.
- Jiao H, Ding M, Zhao H. 2015. Sequence and organization of complete mitochondrial genome of the firefly, Aquatica leii (Coleoptera: Lampyridae). Mitochondrial DNA. 26(5):775–776.
- Kang TH. 2012. Cantharioids I (Arthropoda, Insecta, Coleoptera). Insect fauna of Korea. Volume 12. Incheon: National Institute of Biological Resources.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701.
- Lanfear R, Calcott B, Kainer D, Mayer C, Stamatakis A. 2014. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol Biol. 14:82
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol E. 34:772–773.
- Luan X, Fu X. 2016. The complete mitochondrial genome of the firefly, Asymmetricata circumdata (Motschulsky)(Coleoptera: Lampyridae). Mitochondrial DNA Part B. 1(1):553–555.
- Maeda J, Kato DI, Arima K, Ito Y, Toyoda A, Noguchi H. 2017a. The complete mitogenome and phylogenetic analysis of Japanese firefly ‘Genji Botaru’ Luciola cruciata (Coleoptera: Lampyridae). Mitochondrial DNA Part B. 2(2):522–523.
- Maeda J, Kato DI, Arima K, Ito Y, Toyoda A, Noguchi H. 2017b. The complete mitochondrial genome sequence and phylogenetic analysis of Luciola lateralis, one of the most famous firefly in Japan (Coleoptera: Lampyridae). Mitochondrial DNA Part B. 2(2):546–547.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the 9th Gateway Computing Environments Workshop (GCE), New Orleans. p. 1–8.
- Mu FJ, Ao L, Zhao HB, Wang K. 2016. Characterization of the complete mitochondrial genome of the firefly, Luciola substriata (Coleoptera: Lampyridae). Mitochondrial DNA Part A. 27(5):3360–3362.
- Sriboonlert A, Wonnapinij P. 2019. Comparative mitochondrial genome analysis of the firefly, Inflata indica (Coleoptera: Lampyridae) and the first evidence of heteroplasmy in fireflies. Int J Biol Macromol. 121:671–4676.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Stanger-Hall KF, Lloyd JE, Hillis DM. 2007. Phylogeny of North American fireflies (Coleoptera: Lampyridae): implications for the evolution of light signals. Mol Phylogenet Evol. 45(1):33–49.
- Wang K, Hong W, Jiao H, Zhao H. 2017. Transcriptome sequencing and phylogenetic analysis of four species of luminescent beetles. Sci Rep. 7(1):1814.
