Abstract
Here, we report the first complete mitochondrial genome for the smalltooth sand tiger shark, Odontaspis ferox (Risso, 1810). The circular mitochondrial genome was found to be 16,682 bp in length and contains 37 genes, a control region and the replication origin of the L-strand (OL). The base composition of this mitogenome is 32.6% A, 23.3% C, 12.8% G, and 31.3% T. Phylogenetic analysis of Lamniformes indicates that O. ferox did not group with Carcharias taurus and so the taxonomic classification of Odontaspididae needs to be revised. This study promotes conservation genetics for this poorly studied shark species which is listed critically endangered in the Mediterranean Sea.
The smalltooth sand tiger shark, Odontaspis ferox (Risso, 1810), is one of the most poorly studied shark species (Fergusson et al. Citation2007), which is sparsely distributed in warm-temperate and tropical waters and is considered as uncommon given that it is rarely caught (Compagno Citation2002; Fergusson et al. Citation2007). Through the use of better data collection systems, recent new records of this species’ occurrence are giving a more comprehensive picture of its distribution (White Citation2007; Santander-Neto et al. Citation2011; Acuna-Marrero et al. Citation2013; Ritter and Compagno Citation2013; Long et al. Citation2014; Estupinan-Montano et al. Citation2016; Wellington et al. Citation2017). Moreover, on landing, this species is occasionally misidentified as Hexanchus griseus, given that both species have similar coloration (pers. obs.). Populations of O. ferox are declining and the species has been listed by IUCN as vulnerable on a global scale (Graham et al. Citation2016) and critically endangered at both European (Pollard et al. Citation2015) and Mediterranean level (Pollard et al. Citation2016). Consequently, it has been included in Annex II of the Specially Protected Areas and Biological Diversity (SPA/BD) Protocol (UNEP/MAP-SPA/RAC Citation2018) and in 2012 through the adoption of Recommendation GFCM36/2012/3, General Fisheries Commission for the Mediterranean prohibited the possession and commercialization of this species, while emphasizing on its possible unharmed release (FAO Citation2012). Subsequently, a number of Mediterranean countries have listed O. ferox as a protected species.
A 264 cm O. ferox male specimen was by-caught on 1 February 2011 through trawling activities in Maltese waters by local fishermen (36°5′3″N 14°4′43″E Central Mediterranean Sea). A tissue sample was collected from this specimen as part of fisheries landings sampling undertaken since 2002 by the Conservation Biology Research Group, University of Malta (CBRG-UM). The tissue sample collected for this species was stored at the Ichthyological Collection of the CBRG-UM (Ofer002-110201001) and has already contributed to DNA barcoding of the species (Vella et al. Citation2017). The total genomic DNA was extracted using GF-1 DNA Extraction Kit (Vivantis Technologies, Subang Jaya, Malaysia). A DNA library of the whole genome was constructed and next-generation sequencing reads were generated through Illumina HiSeqX using 2 × 150 bp end reads (Illumina, San Diego, CA). Sequences were paired, trimmed at Q ≥ Q30 and reads shorter that 100 nucleotides were discarded. The final data set was de novo assembled using Geneious R10 (Kearse et al. Citation2012). NCBI ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/) was used to identify PCGs, which were subsequently checked for start and stop codons. The tRNA genes were identified through their secondary structures using tRNAscan-SE v2.0 (Chan and Lowe Citation2019). This newly annotated genome was validated against the mitogenome of other Lamniformes species.
The complete mitogenome for this species is 16,682 bp long (GenBank accession: MT702386) and contains 13 PCGs, two rRNA genes, 22 tRNA genes, and two non-coding regions (control region and OL). The gene order followed the typical vertebrate order (Satoh et al. Citation2016), that is most of the mitochondrial genes are encoded on the H-strand, except one PCG (ND6), eight tRNA genes (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, Pro) and the OL which are encoded on the L-strand. The PCGs range between 168 bp (ATP8) and 1830 bp (ND5) encoding for a total of 3798 amino acids. All PCGs utilize ATG as their start codon except COX1 which uses GTG. The most common stop codon is TAA, while ND6 uses AGG and four genes (COX2, ND3, ND4, and cytB) use T––. The length of the 22 tRNA genes range from 67 bp (SerAGY, Cys) to 75 bp (LeuUUR). All tRNA genes produced the expected cloverleaf structure except for SerAGY that has a missing dihydrouridine arm, typical for most vertebrate species (Satoh et al. Citation2016).
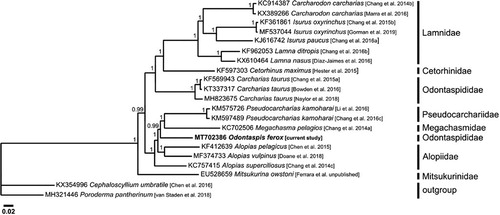
The currently sequenced mitogenome was aligned with that of 13 other Lamniformes using ClustalW (Thompson et al. Citation1994), while a phylogenetic tree, excluding the control region, was constructed using Bayesian Inference analysis through Mr Bayes v3.2.6 (Huelsenbeck and Ronquist Citation2001) () using GTR G + I substitution model as determined by jModelTest v2.1.7 (Darriba et al. Citation2012). This analysis did not group O. ferox with Carcharias taurus even though taxonomic keys place them both in Odontaspididae (Compagno Citation2002). Therefore, current results corroborate a number of morphological studies (Shimada Citation2005; Shimada et al. Citation2009) and molecular phylogenetic studies using smaller DNA sequences (Martin et al. Citation2002; Human et al. Citation2006; Velez-Zuazo and Agnarsson Citation2011; Naylor et al. Citation2012), which show that Odontaspididae is not monophyletic. This outcome indicates that the taxonomic classification of the two species that compose the family Odontaspididae needs to be revised. The genetic resources made available by this study on O. ferox aid future research into the genetics and evolution of its populations. It also promotes knowledge and research to better understand this species’ taxonomy to target effective measures toward its urgent conservation needs.
Figure 1. Bayesian inference based phylogeny depicting the mitogenomic relationship between 14 Lamniformes species using two Carcharhiniformes species as outgroup as inferred from their complete mitogenomes (excluding the control region). Each label includes the GenBank accession number, species, reference and respective family, while numbers at the nodes indicate the posterior probability values. This analysis used 5 × 106 generations, a sample frequency every 1000 generations and a burn-in of 25%. The mean standard deviation of split frequencies was <0.001.
Ethical approval
This study did not require ethical approval as it made use of muscle tissue collected from a dead specimen that was caught by a local fishermen and was sold at the local fish market.
Acknowledgements
The authors would like to thank Maltese fishermen and the Ministry for Sustainable Development, the Environment and Climate Change for supporting this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank (accession no. MT702386) at https://www.ncbi.nlm.nih.gov/genbank/.
Additional information
Funding
References
- Acuna-Marrero D, Zimmerhackel JS, Mayorga J, Hearn A. 2013. First record of three shark species, Odontaspis ferox, Mustelus albipinnis and Centrophorus squamosus, from the Galápagos Islands. Mar Biodivers Rec. 6:e87.
- Bowden DL, Vargas-Caro C, Ovenden JR, Bennett MB, Bustamante C. 2016. The phylogenomic position of the grey nurse shark Carcharias taurus Rafinesque, 1810 (Lamniformes, Odontaspididae) inferred from the mitochondrial genome. Mitochondrial DNA Part A. 27(6):4328–4330.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Chang CH, Chiang WC, Lin YS, Jang-Liaw NH, Shao KT. 2016a. Complete mitochondrial genome of the longfin mako shark, Isurus paucus (Chondrichthyes, Lamnidae). Mitochondrial DNA Part A. 27(1):690–691.
- Chang CH, Jabado RW, Lin YS, Shao KT. 2015a. The complete mitochondrial genome of the sand tiger shark, Carcharias taurus (Chondrichthyes, Odontaspididae). Mitochondrial DNA Part A. 26(5):728–729.
- Chang CH, Jang-Liaw NH, Lin YS, Carlisle A, Hsu HH, Liao YC, Shao KT. 2016b. The complete mitochondrial genome of the salmon shark, Lamna ditropis (Chondrichthyes, Lamnidae). Mitochondrial DNA Part A. 27(1):316–317.
- Chang CH, Shao KT, Lin YS, Chiang WC, Jang-Liaw NH. 2014a. Complete mitochondrial genome of the megamouth shark Megachasma pelagios (Chondrichthyes, Megachasmidae). Mitochondrial DNA. 25(3):185–187.
- Chang CH, Shao KT, Lin YS, Fang YC, Ho HC. 2014b. The complete mitochondrial genome of the great white shark, Carcharodon carcharias (Chondrichthyes, Lamnidae). Mitochondrial DNA. 25(5):357–358.
- Chang CH, Shao KT, Lin YS, Ho HC. 2016c. The complete mitochondrial genome of the crocodile shark, Pseudocarcharias kamoharai (Chondrichthyes, Lamnidae). Mitochondrial DNA Part A. 27(3):1948–1949.
- Chang CH, Shao KT, Lin YS, Ho HC, Liao YC. 2014c. The complete mitochondrial genome of the big-eye thresher shark, Alopias superciliosus (Chondrichthyes, Alopiidae). Mitochondrial DNA. 25(4):290–292.
- Chang CH, Shao KT, Lin YS, Tsai AY, Su PX, Ho HC. 2015b. The complete mitochondrial genome of the shortfin mako, Isurus oxyrinchus (Chondrichthyes, Lamnidae). Mitochondrial DNA. 26(3):475–476.
- Chen H, Lin L, Chen X, Ai W, Chen S. 2016. Complete mitochondrial genome and the phylogenetic position of the Blotchy swell shark Cephaloscyllium umbratile. Mitochondrial DNA Part A. 27(4):3045–3047.
- Chen X, Xiang D, Ai W, Shi X. 2015. Complete mitochondrial genome of the pelagic thresher Alopias pelagicus (Lamniformes: Alopiidae). Mitochondrial DNA. 26(2):323–324.
- Compagno LJV. 2002. Sharks of the World: an annotated and illustrated catalogue of shark species known to date. Volume 2. Bullhead, mackerel and carpet sharks (Heterodontiformes, Lamniformes and Orectolobiformes). FAO species catalogue for fishery purposes. Rome: FAO.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Diaz-Jaimes P, Uribe-Alcocer M, Adams DH, Rangel-Morales JM, Bayona-Vásquez NJ. 2016. Complete mitochondrial genome of the porbeagle shark, Lamna nasus (Chondrichthyes, Lamnidae). Mitochondrial DNA Part B. 1(1):730–731.
- Doane MP, Kacev D, Harrington S, Levi K, Pande D, Vega A, Dinsdale EA. 2018. Mitochondrial recovery from shotgun metagenome sequencing enabling phylogenetic analysis of the common thresher shark (Alopias vulpinus). Meta Gene. 15:10–15.
- Estupinan-Montano C, Galvan-Magana F, Hacohen-Domene A, Estupinan-Ortiz JF. 2016. First reports of smalltooth sand tiger sharks, Odontaspis ferox (Elasmobranchii: Lamniformes: Odontaspididae), off the continental Ecuador. Acta Ichthyol Piscat. 46:251–253.
- FAO. 2012. Recommendation GFCM/36/2012/3 on fisheries management measures for conservation of sharks and rays in the GFCM area. http://www.fao.org/3/a-ax385e.pdf.
- Fergusson IK, Graham KJ, Compagno LJV. 2007. Distribution, abundance, and biology of the smalltooth sandtiger Odontaspis ferox (Risso, 1810) (Lamniformes: Odontaspididae). Environ Biol Fish. 81(2):207–228.
- Gorman J, Marra N, Shivji MS, Stanhope MJ. 2019. The complete mitochondrial genome of an Atlantic Ocean Shortfin Mako Shark, Isurus oxyrinchus. Mitochondrial DNA B Resour. 4(2):3642–3643.
- Graham KJ, Pollard DA, Gordon I, Williams S, Flaherty AA, Fergusson I, Dicken M. 2016. Odontaspis ferox. The IUCN red list of threatened species 2016. e.T41876A2957320.en.
- Hester J, Atwater K, Bernard A, Francis M, Shivji MS. 2015. The complete mitochondrial genome of the basking shark Cetorhinus maximus (Chondrichthyes, Cetorhinidae). Mitochondrial DNA. 26(5):730–731.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Human BA, Owen EP, Compagno LJV, Harley EH. 2006. Testing morphologically based phylogenetic theories within the cartilaginous fishes with molecular data, with special reference to the catshark family (Chondrichthyes; Scyliorhinidae) and the interrelationships within them. Mol Phylogenet Evol. 39(2):384–391.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li W, Dai X, Tian S, Xu Q, Wu F, Gao C, Zhang Y. 2016. Complete mitochondrial genome of the crocodile shark Pseudocarcharias kamoharai (Lamniformes: Pseudocarchariidae). Mitochondrial DNA Part A. 27(3):2095–2097.
- Long DJ, Sala E, Ballesteros E, Caselle JE, Friedlander AM, Klapfer A, Blum S, Constable HB. 2014. Summary of South American records of the smalltooth sand tiger shark Odontaspis ferox (Chondrichthyes: Odontaspidae), with the first record from Chilean waters. Marine Biodivers Rec. 7:e67.
- Marra NJ, Wang M, Sun Q, Pavinski Bitar PD, Stanhope MJ, Shivji MS. 2016. Mitochondrial genome of an Atlantic white shark (Carcharodon carcharias). Mitochondrial DNA Part B. 1(1):717–719.
- Martin AP, Pardini AT, Noble LR, Jones CS. 2002. Conservation of a dinucleotide simple sequence repeat locus in sharks. Mol Phylogenetics Evol. 23(2):205–213.
- Naylor GJP, Caira JN, Jensen K, Rosana KAM, White WT, Last PR. 2012. A DNA sequence-based approach to the identification of shark and ray species and its implications for global Elasmobranch diversity and parasitology. Bull Am Mus Nat Hist. 367:1–263.
- Naylor GJP, Yang L, Corrigan SL, French L. 2018. New York shark bites: DNA result should calm the waters. Nature. 561(7721):33.
- Pollard DA, Gordon I, Williams S, Flaherty AA, Fergusson I, Dicken M, Graham K. 2015. Odontaspis ferox. The IUCN red list of threatened species 2015. e.T41876A48949693.
- Pollard DA, Gordon I, Williams S, Flaherty AA, Fergusson I, Dicken M, Graham K. 2016. Odontaspis ferox. The IUCN red list of threatened species 2016. e.T41876A16527837.
- Ritter EK, Compagno LJV. 2013. First record of a smalltooth sandtiger shark, Odontaspis ferox, from the Galápagos Islands. Marine Biodivers Rec. 6:e130.
- Santander-Neto J, Faria VV, Castro ALF, Burgess GH. 2011. New record of the rare ragged-tooth shark, Odontaspis ferox (Chondrichthyes: Odontaspidae) from the south-west Atlantic identified using DNA bar coding. Marine Biodivers Rec. 4:e75.
- Satoh TP, Miya M, Mabuchi K, Nishida M. 2016. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 17:719.
- Shimada K. 2005. Phylogeny of lamniform sharks (Chondrichthyes: Elasmobranchii) and the contribution of dental characters to lamniform systematics. Paleontol Res. 9(1):55–72.
- Shimada K, Rigsby CK, Kim SH. 2009. Labial cartilages in the smalltooth sandtiger shark, Odontaspis ferox (Lamniformes: Odontaspididae) and their significance to the phylogeny of Lamniform Sharks. Anat Rec (Hoboken). 292(6):813–817.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 22:4673–4680.
- UNEP/MAP-SPA/RAC. 2018. SAP/RAC:SPA-BD protocol – annex II: list of endangered or threatened species. https://www.rac-spa.org/sites/default/files/annex/annex_2_en_20182.pdf.
- van Staden M, Gledhill KS, Rhode C, Bester-van der Merwe AE. 2018. The complete mitochondrial genome and phylogenetic position of the leopard catshark, Poroderma pantherinum. Mitochondrial DNA Part B. 3(2):750–752.
- Velez-Zuazo X, Agnarsson I. 2011. Shark tales: a molecular species-level phylogeny of sharks (Selachimorpha, Chondrichthyes). Mol Phylogenet Evol. 58(2):207–217.
- Vella A, Vella N, Schembri S. 2017. A molecular approach towards taxonomic identification of elasmobranch species from Maltese fisheries landings. Mar Genomics. 36:17–23.
- Wellington CM, Wakefield CB, White WT. 2017. First record of Odontaspis ferox (Risso, 1810) in the temperate south-eastern Indian Ocean from in situ observations in a deep-water canyon using baited video. J Appl Ichthyol. 33(1):133–135.
- White W. 2007. Biological observations on lamnoid sharks (Lamniformes) caught by fisheries in eastern Indonesia. J Mar Biol Ass. 87(3):781–788.

