Abstract
Donkey croaker, Pennahia anea (Bloch, 1793) is a commercially important croaker in the Indo-Pacific region. In this study, we sequenced and analyzed the mitogenome of P. anea. The nearly complete mitochondrial genome of P. anea is 15,694 bp in size. It contained 13 protein-coding genes (PCGs), 2 rRNA genes, and 22 tRNA genes. The sequence had the A-T content of 55.4% and GC content of 44.6%. All 13 PCGs used ATG codon for initiation, while TAA codon was the most common for termination. Phylogenetic analysis demonstrated that P. anea is located within the genus Pennahia. This study provides additional data for the understanding of the phylogeny of the family Sciaenidae.
Keywords:
Sciaenidae is a family of the order Perciformes that contains around 300 species worldwide (Quan et al. Citation2011; Froese and Pauly Citation2019). Some species of this family are used as food fish and are important targets for commercial fishery. The ability to produce a croaking sound is known as a distinguishing characteristic of sciaenids (Ramcharitar et al. Citation2006). Pennahia anea is one of five species of the genus Pennahia, a member of the family Sciaenidae. This species is distributed in the Indo-Pacific region, including Vietnamese waters (Froese and Pauly Citation2019). P. anea is a commercially important species in the distributed region. Of five species of the genus Pennahia, three have been sequenced mitogenomes, namely Pennahia argentata, Pennahia macrocephalus, and Pennahia pawak. Sequencing the mitogenome of the remaining species is necessary to understand phylogenetic relationships within the genus.
Specimens of P. anea were collected from Phu Long - Cat Hai, Vietnam (20°45'15.06"N - 106°54'53.62"E) during November 2019 to August 2020. A specimen used for mitogenome sequencing was deposited at the Institute of Marine Environment and Resources, Vietnam (contact person: Chien Pham Van, email: [email protected]) under the voucher number IMER-TA-007. Following DNA extraction, 150 bp paired-end libraries were prepared and analyzed using the MGISEQ-2000 system (MGI, Shenzen, China). Subsequently, raw data were assembled using Mitobim program and annotation was performed using MITOS program (Bernt et al. Citation2013; Hahn et al. Citation2013). A phylogenetic tree of the family Sciaenidae was constructed based on 13 protein-coding genes (PCGs). To build the tree, Bayesian inference method in MrBayes version 3.2.7a was applied with two independent runs, 10 million generations, and sampling every 1000 generations (Ronquist et al. Citation2012).
Through the sequencing, the nearly complete mitogenome of P. anea was obtained. The nearly complete mitogenome of P. anea (GeneBank accession number: MW408700) was 15,694 bp long and contained the typical set of 37 genes, including 13 PCGs, and 2rRNA genes, and 22 tRNA genes. The nucleotide composition of the sequence was 27.3% A, 28.2% C, 16.4% G and 28.1% T. Base composition is similar to that observed in other Perciformes species (Ceruso, Mascolo, Lowe, et al. Citation2018, Ceruso, Mascolo, Palma, et al. Citation2018; Mascolo et al. Citation2018a, Citation2018b, Citation2019; Ceruso et al. Citation2020). The sizes of genes in the mitogenome of P. anea were comparable to that of other Pennahia species (Li et al. Citation2014; Guo et al. Citation2016; Lin et al. Citation2017). The sizes of PCGs ranged from 168 bp (atp8) to 1,815 bp (nd5) and the sizes of tRNA genes ranged from 68 bp (tRNA-Ser) to 75 bp (tRNA-Lys and tRNA-Leu). Two rRNA genes, 12S rRNA and 16S rRNA were 958 and 1,709 bp in size. All PCGs used ATG codon for initiation. Meanwhile, five of 13 PCGs used TAA codon for termination (atp6, atp8, nd1, nd4l, and nd6). AGA termination codon was observed in cox1 and nd4, and TAG termination codon was observed in nd5 while AGG termination codon was observed in cytb and nd3. The incomplete termination codon was found in cox2, cox3, and nd2 (Miya et al Citation2001; Ceruso et al. Citation2019).
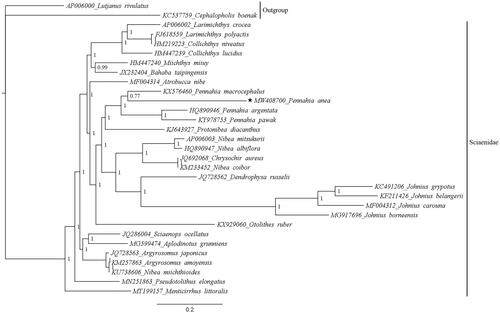
Phylogenetic analysis based on nucleotide sequences of 13 PCGs showed the position of P. anea in the family Sciaenidae (). The phylogenetic tree revealed that P. anea was clustered with other members of the genus Pennahia, which was sister to Protonibea diacanthus. Our finding is in agreement with previous studies that showed the relationships of the genus Pennahia with other species of the family Sciaenidae (Lin et al. Citation2017; Wen et al. Citation2020). The mitogenome reported from this study might be helpful for future studies that deal with the genetic diversity of P. anea.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study is openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW408700. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA694527, SRR13555243, and SAMN17376889, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Ceruso C, Venuti I, Osca D, Caputi L, Anastasio A, Crocetta F, Sordino P, Pepe T. 2020. The complete mitochondrial genome of the sharpsnout seabream Diplodus puntazzo (Perciformes: Sparidae). Mitochondrial DNA Part B. 5(3):2379–2381.
- Ceruso M, Mascolo C, Lowe EK, Palma G, Anastasio A, Pepe T, Sordino P. 2018. The complete mitochondrial genome of the common Pandora Pagellus erythrinus (Perciformes: Sparidae). Mitochondrial DNA Part B. 3(2):624–625.
- Ceruso M, Mascolo C, Palma G, Anastasio A, Pepe T, Sordino P. 2018. The complete mitochondrial genome of the common dentex, Dentex dentex (Perciformes: Sparidae). Mitochondrial DNA Part B. 3(1):391–392.
- Ceruso M, Mascolo C, Anastasio A, Pepe T, Sordino P. 2019. Frauds and fish species authentication: study of the complete mitochondrial genome of some sparidae to provide specific barcode markers. Food Control. 103:36–47.
- Froese R, Pauly D. 2019. FishBase. [accessed December 05, 2020]. www.fishbase.org.
- Guo CC, Liu M, Lin JJ, Jiang XB. 2016. Complete mitochondrial genome and the phylogenetic position of the bighead pennah croaker Pennahia macrocephalus (Perciformes: Sciaenidae). Mitochondrial DNA Part B. 1(1):759–760.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Li A, Sun D, Li Y, Gao T. 2014. Mitochondrial genome sequence of the Japanese white croaker Pennahia argentata (Perciformes: Sciaenidae). Mitochondrial DNA. 25(2):117–119.
- Lin BA, Guo CC, Liu M. 2017. Complete mitochondrial genome and the phylogenetic position of the Pawak croaker Pennahia pawak (Perciformes: Sciaenidae). Mitochondrial DNA Part B. 2(1):366–368.
- Mascolo C, Ceruso M, Chirollo C, Palma G, Anastasio A, Sordino P, Pepe T. 2019. The complete mitochondrial genome of the Angolan dentex Dentex angolensis (Perciformes: Sparidae). Mitochondrial DNA Part B. 4(1):1245–1246.
- Mascolo C, Ceruso M, Palma G, Anastasio A, Sordino P, Pepe T. 2018a. The complete mitochondrial genome of the axillary seabream, Pagellus acarne (Perciformes: Sparidae). Mitochondrial DNA Part B. 3(1):434–435.
- Mascolo C, Ceruso M, Palma G, Anastasio A, Sordino P, Pepe T. 2018b. The complete mitochondrial genome of the Pink dentex Dentex gibbosus (Perciformes: Sparidae). Mitochondrial DNA Part B. 3(2):525–526.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18(11):1993–2009.
- Quan VN, Chien PV, The ND. 2011. The species composition of the croaker fish (family Sciaenidae) in the coastal zone of Quang Ninh and Hai Phong province. Proceedings of the Workshop “Coastal Marine Biodiversity and Bioresources of Vietnam and Adjacent Areas to the South China Sea”, Nha Trang, Vietnam; p. 90–94.
- Ramcharitar J, Gannon DP, Popper AN. 2006. Bioacoustics of fishes of the family Sciaenidae. Trans Am Fish Soc. 135(5):1409–1431.
- Ronquist F, Teslenko M, van der Mark P, Ayres D, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Wen HB, Ma X, Xu P, Cao Z, Zheng B, Hua D, Jin W, Sun G, Gu R. 2020. Structure and evolution of the complete mitochondrial genome of the freshwater drum, Aplodinotus grunniens (Actinopterygii: Perciformes: Sciaenidae). Acta Ichthyol Piscat. 50(1):23–35.