Abstract
The complete chloroplast (cp) genome of Ardisia brevicaulis Diels 1900, one traditionally medicinal plant usually used in southern China, was first assembled and reported in this study. The genome size is 156,742 bp (37.1% GC content), containing a large single-copy (LSC) region of 86,329 bp, a small single-copy (SSC) region of 18,417 bp, and a pair of inverted repeat regions (IRs) of 25,998 bp. 134 genes (89 protein-coding, 37 tRNA, and 8 rRNA genes) are annotated in the whole cp genome, including 115 unique genes (81 protein-coding, 30 tRNA, and 4 rRNA genes). Phylogenetic analysis showed that A. brevicaulis is closely related to A. primulifolia and A. villosa, indicating their close phylogenetic relationship. The cp genome of A. brevicaulis could provide valuable genomic information for the phylogeny, molecular identification and discovery of new medicinal plant resources in Ardisia Swartz 1788.
Ardisia brevicaulis Diels 1900, one species of Ardisia Swartz 1788 from the family Primulaceae Batsch ex Borkh.1797 (Chen and Pipoly Citation1996; APG IV Citation2016), is an important medicinal plant species widely distributed in southern China (González de Mejía and Ramírez-Mares Citation2011). The root of A. brevicaulis is usually used as a traditional chinese medicine for menstrual disorders, injuries and rheumatic pains (González de Mejía and Ramírez-Mares Citation2011; Li et al. Citation2010). Moreover, chemical and pharmacological studies showed that abundant natural products isolated from the root of A. brevicaulis have the potential biological activity of anti-tumor effect (Chen et al. Citation2011; Zhu et al. Citation2012). In particular, Ardisiphenol D, a major active resorcinol derivative among all the products, was shown to have potent inhibitory activity against cancer cells both in vitro and in vivo (Zhao et al. Citation2014). Due to the large number of species (65 species in China and about 400 to 500 species worldwide) in Ardisia and the similar morphology of Ardisia species (Chen and Pipoly Citation1996), the morphological classification and identification within the genus are difficult (Julius et al. Citation2021). Molecular phylogeny based on nrITS better revealed that Ardisia is not monophyletic as currently circumscribed, but left the phylogenetic relationship among many species of Ardisia unresolved, including the relationship between A. brevicaulis and its related species (Julius et al. Citation2021). In molecular phylogenetics, the chloroplast genomes can provide valuable sources of phylogenetic information because of their relatively stable genome structure and high evolutionary rates (Ravi et al. Citation2008). In this study, we first reported the complete chloroplast (cp) genome of A. brevicaulis and constructed the phylogenetic relationship with other Ardisia species. The results could be helpful for the phylogeny, molecular identification and discovery of new medicinal plant resources in Ardisia.
Materials
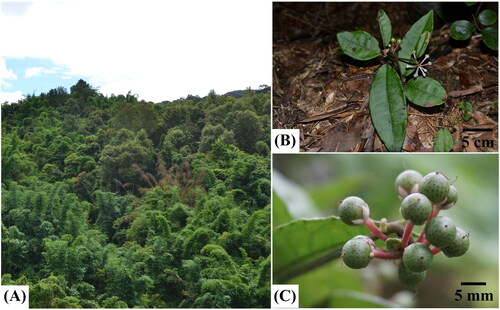
The sample of A. brevicaulis was collected from a wild population (108°35′12.12″E, 25°34′52.32 N; 910 m) in Congjiang County, Guizhou Province, China. It is a subshrub (10–15 cm) usually growing in dark and humid places under mixed forests (; Chen and Pipoly Citation1996). It is characterized by its narrowly ovate to elliptic or suboblong leaf blade (7–18 × 2.5–6 cm) with an entire margin and indistinct marginal glandular dots, terminal umbellate inflorescences, globose punctate fruits (c. 6 mm in diameter) with usually purplish red persistent calyx and fruiting stalk (; Chen and Pipoly Citation1996). The specimen of A. brevicaulis in this study was deposited at the herbarium of Guizhou University of Traditional Chinese Medicine (Chenggang Hu, Email: [email protected]) under voucher number WZW20210304.
Methods
Genomic DNA was extracted from fresh leaves of A. brevicaulis using Tiangen DNA Extraction Kit (DP305, Tiangen, Beijing, China) following the instructions of the kit. Purified genomic DNA was sheared into fragments (c. 350 bp) to construct a paired-end (PE) library. PE reads (c. 150 bp) were generated based on Illumina NovaSeq 6000 (Beijing Novogene Technology Co. Ltd). In total, 1.14 Gb clean data was retained after removing low quality reads and adaptor sequences. Based on the clean data, we assembled the cp genome of A. brevicaulis using GetOrganelle v1.7.5 (Jin et al. Citation2020), and annotated it via PGA (Qu et al. Citation2019), with further manual check and adjustment in Geneious v10.2.2 (Kearse et al. Citation2012). Annotated cp genome of A. brevicaulis was deposited to NCBI under the accession number ON208988.
The phylogenetic relationship between A. brevicaulis and other Ardisia species was inferred based on the whole cp genome sequences, with Tapeinosperma multiflorum and T. netor as outgroups (Hou et al. Citation2019; Shi and Liu Citation2019; Yan et al. Citation2019). Sequences in this study were aligned using MAFFT v7.397 (Katoh and Standley Citation2013). A maximum-likelihood (ML) tree was reconstructed in IQ-TREE v2.2.0 (Minh et al. Citation2020) using the best-fit substitution model (K3Pu + F+R2) chosen by ModelFinder (Kalyaanamoorthy et al. Citation2017). Branch supports were tested using SH-like approximate likelihood ratio (SH-aLRT) and Ultrafast bootstrap (UFBoot) with 10,000 bootstrap replicates.
Results
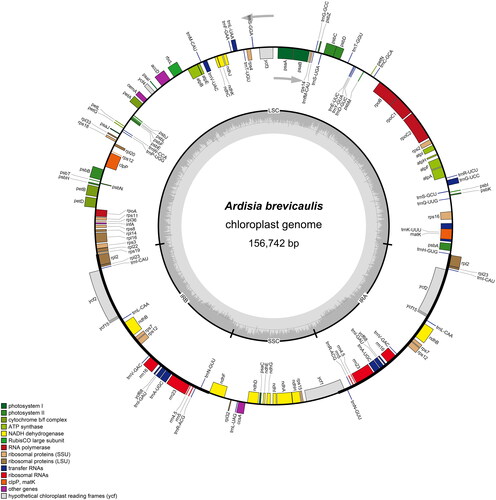
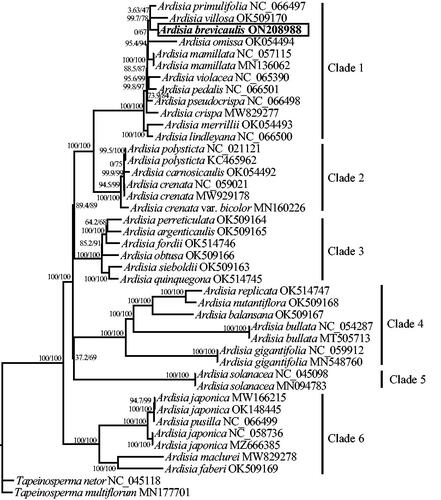
The total length of the cp genome is 156,742 bp, with a total GC content of 37.1%. A typical quadripartite structure was shown in the cp genome, including a large single-copy (LSC) region (86,329 bp), a small single-copy (SSC) region (18,417 bp), and two inverted repeat regions (IRs; 25,998 bp). 134 genes (89 protein-coding, 37 tRNA, and 8 rRNA genes) were annotated in the whole cp genome, containing 115 unique genes (81 protein-coding, 30 tRNA, and 4 rRNA genes). Among them, 15 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) contained one intron, while the clpP and ycf3 genes contained two introns. In addition, rps12 gene underwent trans-splicing (). The phylogenetic tree in this study revealed that A. brevicaulis is nested in one well-supported clade (Clade1; 100/100%) with other 10 Ardisia species, in which A. brevicaulis is closely related to A. primulifolia and A. villosa (99.7/78%; ).
Discussion and conclusion
Ardisia is the largest genus from the family Primulaceae and one of the largest tropical genera with an estimate of about 400 to 500 species mainly distributed in the tropical Asia, Americas, Australia, and the Pacific Islands (APG IV Citation2016; Chen and Pipoly Citation1996; Frodin Citation2004). Its classification has received considerable attention from botanists in decades, but mainly focused on morphological classification, and presented great uncertainties (Julius et al. Citation2021). Recently, a molecular phylogeny of Ardisia based on nrITS and three cpDNA regions suggested the monophyly of subgenera Akosmos, Bladhia and Crispardisia, and the non-monophyly of subgenera Tinus and Icacorea, but cannot distinguish many species within Ardisia well (Ku and Hu Citation2014). Julius et al. (Citation2021) explored a comprehensive phylogenetic relationships among Ardisia and related genera from 177 samples based on nrITS. It well indicate the non-monophyly of Ardisia, but clearly suggested the need for further study of the phylogenetic relationship within Ardisia because of low supporting rates among many Ardisia species. In this study, the phylogenetic relationship among surveyed Ardisia species was well resolved, showing six well-supported clades (). Among them, A. brevicaulis and other 10 Ardisia species were nested in one clade with high supporting rates (100/100%), in which A. brevicaulis is closely related to A. primulifolia and A. villosa (). These results probably implied that the chloroplast genomes can better resolve the interspecific relationship within Ardisia. Moreover, since medicinal plants within the same phylogenetic groups may have the same or similar therapeutic compounds/effects (Hao and Xiao Citation2020; Gong et al. Citation2022;), it is likely that the phylogenetic results in this study can also provide useful information for the exploration of new medicinal resources in Ardisia. However, it is worth noting that our phylogenetic tree was constructed only based on 30 Ardisia species (including one variety) because of few reports and releases of the chloroplast genomes of Ardisia in NCBI at present. Given the numerous species and wide distribution of Ardisia, in future studies, more chloroplast genomes and more species of Ardisia from all over the world are badly needed to be sequenced and studied to better explore the evolution and phylogenetic relationship within the genus.
Ethical approval
Ardisia brevicaulis Diels 1900 is not protected nor endangered in China. Research and collection of plant material comply with appendices I, II and III of the Convention on the Trade in Endangered Species of Wild Fauna and Flora, which has been valid from 22 June 2022 (https://cites.org/eng/app/appendices.php).
Author contributions
Zhiwei Wang analyzed the data and wrote the paper. Youqiong Hu performed the experiments. Shenghua Wei conceived and designed the study. All authors have read and approved the manuscript for publication, and all authors agreed to be accountable for all aspects of the work.
Acknowledgments
The authors are very grateful to Anling Huang of Guizhou University of Traditional Chinese Medicine for her help in collecting the experimental samples.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data in this study is openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession number ON208988. The associated BioProject, SRA, and Bio-Sample numbers involved in this study are PRJNA824992, SRR18700737, and SAMN27502973 respectively.
Additional information
Funding
References
- APG IV. 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc. 181(1):1–20.
- Chen J, Pipoly JJ. 1996. Myrsinaceae. In Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol.15. Beijing: Science Press; Saint Louis: Missouri Botanical Garden Press; p. 1–38.
- Chen L-P, Zhao F, Wang Y, Zhao L-L, Li Q-P, Liu H-W. 2011. Antitumor effect of resorcinol derivatives from the roots of Ardisia brevicaulis by inducing apoptosis. J Asian Nat Prod Res. 13(8):734–743.
- Frodin DG. 2004. History and concepts of big plant genera. Taxon. 53(3):753–776.
- Gong X, Yang M, He C, Bi Y, Zhang C, Li M, Xiao P. 2022. Plant Pharmacophylogeny: review and future directions. Chin J Integr Med. 28(6):1–8.
- González de Mejía E, Ramírez-Mares MV. 2011. Ardisia: health-promoting properties and toxicity of phytochemicals and extracts. Toxicol Mech Methods. 21(9):667–674.
- Hao D, Xiao P. 2020. Plant pharmacophylogeny: past, present and future. J Chin Pharm Sci. 29(12):831–854.
- Hou N, Li M, Shen J, Chen Z, Luo Y, Deng L. 2019. The complete chloroplast genome of Ardisia mamillata (Myrsinaceae). Mitochondrial DNA Part B. 4(2):3441–3442.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Julius A, Sabran S, Gutierrez-Ortega JS. 2021. Phylogenetic relationship of tropical Asian Ardisia and relatives (Primulaceae) shows non-monophyly of recognized genera and subgenera. J Jpn Bot. 96(3):149–165.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al.,. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Ku C, Hu JM. 2014. Phylogenetic and cophylogenetic analyses of the leaf-nodule symbiosis in Ardisia subgenus Crispardisia (Myrsinaceae): evidence from nuclear and chloroplast markers and bacterial rrn operons. Int J Plant Sci. 175(1): 92–109.
- Li B, Wang M, Feng Z, Zhao Y, Liu H. 2010. Two new resorcinol derivatives with strong cytotoxicity from the roots of Ardisia brevicaulis Diels. Chem Biodivers. 7(12):2901–2907.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Haeseler A, Lanfear R. 2020. Corrigendum to: IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(8):2461.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Ravi V, Khurana JP, Tyagi AK, Khurana P. 2008. An update on chloroplast genomes. Plant Syst Evol. 271(1-2):101–122.
- Shi Y, Liu B. 2019. Complete chloroplast genome sequence of Ardisia gigantifolia (Myrsinaceae), a vulnerable medicinal plant. Mitochondrial DNA Part B. 4(2):4037–4038.
- Yan X, Liu T, Yuan X, Xu Y, Yan H, Hao G. 2019. Chloroplast genomes and comparative analyses among thirteen taxa within Myrsinaceae s.str. clade (Myrsinoideae, Primulaceae). IJMS. 20(18):4534.
- Zhao F, Hu Y, Chong C, Lu M, Chen L, Kan W, Chen L, Liu H. 2014. Ardisiphenol D, a resorcinol derivative identified from Ardisia brevicaulis, exerts antitumor effect through inducing apoptosis in human non-small-cell lung cancer A549 cells. Pharm Biol. 52(7):797–803.
- Zhu GY, Wong B, Lu A, Bian ZX, Zhang G, Chen HB, Wong YF, Fong WF, Yang Z. 2012. Alkylphenols from the roots of Ardisia brevicaulis induce G1 arrest and apoptosis through endoplasmic reticulum stress pathway in human non-small-cell lung cancer cells. Chem Pharm Bull. 60(8):1029–1036.