Abstract
Berchemia racemosa Siebold & Zucc., 1845 is a rare species distributed in restricted areas in the western Korean peninsula. In this study, the complete chloroplast genome (plastome) of B. racemosa was sequenced and assembled by Illumina paired-end sequencing. The plastome of B. racemosa was 161,187 bp in length and was quadripartite in structure, including a large single-copy (LSC) region of 89,503 bp, a small single-copy (SSC) region of 18,214 bp, and two inverted repeats of 26,735 bp. The GC content was 37.2%. The plastome of B. racemosa contains 130 genes, including eight ribosomal RNA (rRNA) genes, 37 transfer RNA (tRNA) genes, and 85 protein-coding genes. Phylogenetic analysis using complete genome sequences showed that B. racemosa is most closely related to Berchemia flavescens.
Introduction
Berchemia racemosa Siebold & Zucc., 1845 is a deciduous vine in the Rhamnaceae family that is native to Korea, Japan, and Taiwan. In Korea, B. racemosa is a rare plant distributed only in a limited region in Jeollabuk-do Province. Its natural habitat in Jeollabuk-do Province is designated as a nature reserve. Here, we analyzed the complete chloroplast genome (plastome) sequence of B. racemosa to provide valuable information for genetic diversity and phylogenetic relationships between B. racemosa and other species in the Rhamnaceae family.
Materials and methods
Sampling and genome sequencing
Plant materials of B. racemosa were sampled from Gunsan, Jeollabuk-do Province, Korea (126°41′23.10″E, 35°58′41.90″N). Each specimen was imaged using a digital camera to record the information about the sampling sites in natural habitat (). The specimens of B. racemosa were identified by Joon Moh Park and deposited in the Jeollabuk-do Forest Environment Research Institute, South Korea (https://forest.jb.go.kr/; contact person, Joon Moh Park; E-mail, [email protected]) under the voucher number JFERI0020-2. Research plan for sample collection and experimental methods in this study has passed the ethical survey of the Plant Ethics Committee from Jeollabuk-do Forest Environment Research Institute. Total genomic DNA was extracted from the fresh leaf of B. racemosa using an Exgene Plant SV kit (GeneAll Inc., Seoul, South Korea). The DNA was mechanically fragmented to create the sequencing library, and both ends of the DNA were then ligated with adaptors using the TruSeq DNA PCR Free kit (Macrogen Inc., Seoul, South Korea). A paired-end library with an insert size between 350 and 450 bp (Supplementary Tables 1–3) was sequenced using the Illumina HiSeq X platform (Macrogen Inc., Seoul, South Korea).
Assembly and annotation of chloroplast genome
The complete plastome was assembled using NOVOPlasty v.4.3.1 (Dierckxsens et al. Citation2017). Gene annotation was performed using GeSeq v.1.59 (Tillich et al. Citation2017) with options of Chloe v. 0.1.0., BLATN, and BLATX. BLAST was used to further identify positions of inverted repeat (IR) regions by searching against the published plastome database. A circular map of the complete plastome was generated by CPGView software (http://www.1kmpg.cn/cpgview/). The genomic DNA of B. racemosa was deposited in the Jeollabuk-do Forest Environment Research Institute (contact person, Joon Moh Park; E-mail, [email protected]) under the voucher number JFERI-DNA0020-2. All biological samples and research in this study had been approved by the Ethics Committee of Jeollabuk-do Forest Environment Research Institute. To clarify the phylogenetic position of B. racemosa, the complete plastome sequences of 47 Rosales species and two outgroups (Castanea mollissima and Cucurbita moschata) were downloaded from GenBank and aligned using MAFFT v7.3 (Katoh and Standley Citation2013). Spurious matches or poorly aligned regions were removed from the multiple sequence alignment by TrimmAl v.1.2 (Capella-Gutiérrez et al. Citation2009). The resulting aligned sequences of 127,121 bp were analyzed by the maximum-likelihood (ML) method using IQ-Tree (Nguyen et al. Citation2015) with a TVM + F+R4 substitution model as a best-fit model, 1000 replicates of ultrafast bootstrap, and SH-aLRT branch support.
Results
Chloroplast genome features
The complete plastome of B. racemosa (GenBank accession number ON749761) is 161,187 bp with 37.2% GC content. It is composed of a pair of IR regions of 26,735 bp, a large single-copy (LSC) region of 89,503 bp, and a small single-copy (SSC) region of 18,214 bp (). The gene structure of the plastome was nearly identical to that of other Rhamnaceae species (Ma et al. Citation2017). A total of 130 genes were annotated in the plastome, comprising eight ribosomal RNA (rRNA) genes, 37 transfer RNA (tRNA) genes, and 85 protein-coding genes (PCGs). The rps12 is a trans-spliced gene. The 5′ exon of this gene is found in the LSC region, while the 3′ exon is duplicated in the IR regions (). Thirteen genes, including rps16, atpF, rpoC1, pafI, clpP1, petB, petD, rps16, rpl2, ndhB, ndhA, ndhB, and rpl2, contain one or two introns ().
Figure 2. Schematic map of B. racemosa complete chloroplast genome constructed by CPGview (http://www.1kmpg.cn/cpgview/). The map contains six tracks. From the center to outward, the first track shows the dispersed repeats. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
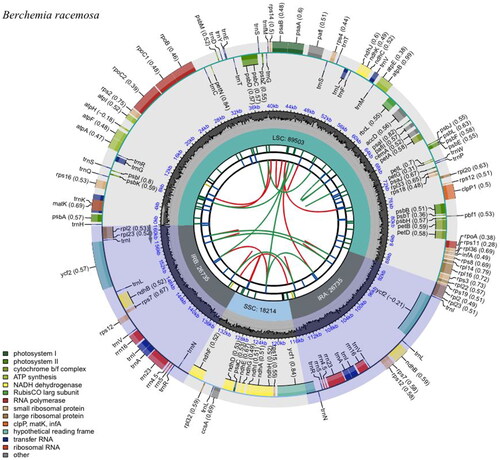
Figure 3. Schematic map of the trans-splicing gene rps12 (A) and cis-splicing genes (B) in the chloroplast genome of B. racemosa. The exons are shown in black; the introns are shown in white. The arrow indicates the sense direction of the gene. The map was generated using CPGview software (http://www.1kmpg.cn/cpgview/).
Whole plastome sequence of seven Rhamnaceae species was aligned to compare the variations of gene structure within the family. The plastome of these species showed high similarity in terms of the number, length, and arrangement of genes. In particular, the plastome sequence of B. racemosa showed the highest sequence similarity of 98.7% with Berchemia flavescens (Zhu et al. Citation2019), except for the inverted orientation of a 13,092-bp fragment from 115,721 to 128,812 bp.
Phylogenetic analysis
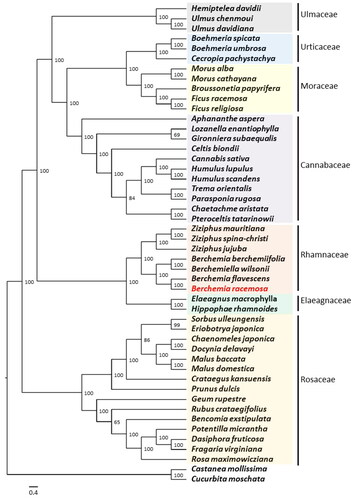
Complete plastome sequences of 47 species from seven families in Order Rosales were utilized to explore the phylogenetic position of B. racemosa. The reconstructed phylogenetic tree showed that B. racemosa is most closely related to Berchemia flavescens (Zhu et al. Citation2019) with high support (BS = 100), both of which belong to the genus Berchemia in the Rhamnaceae family (). It also indicated that Rhamnaceae are monophyletic and sister to the Elaeagnaceae family. In addition, our phylogenetic tree confirmed that all seven families formed monophyletic groups.
Figure 4. Maximum-likelihood (ML) tree showing the relationship among B. racemosa and representative species within Order Rosales based on the complete chloroplast genome sequences. The following sequences were used: Aphananthe aspera (NC_039726; Zhang et al. Citation2018), Bencomia exstipulata (NC_039924), Berchemia berchemiifolia (NC_037477; Cheon et al. Citation2018), Berchemia flavescens (MK460212; Zhu et al. Citation2019), Berchemia racemosa (ON749761; this study), Berchemiella wilsonii (KY926621; Wang et al. Citation2018), Boehmeria spicata (NC_036989; Huang et al. Citation2019), Boehmeria umbrosa (NC_036990; Huang et al. Citation2019), Broussonetia papyrifera (KX828844), Cannabis sativa (KR363961; Oh et al. Citation2016), Cecropia pachystachya (NC_039763; Wu et al. Citation2017), Celtis biondii (NC_039727; Zhang et al. Citation2018), Chaenomeles japonica (KT932966), Chaetachme aristata (MH118120; Zhang et al. Citation2018), Crataegus kansuensis (MF784433; Zhang et al. Citation2020), Dasiphora fruticosa (MF683841; Zhao et al. Citation2018), Docynia delavayi (KX499860; Zhang et al. Citation2017), Elaeagnus macrophylla (KP211788; Choi et al. Citation2015), Eriobotrya japonica (KT633951; Shen et al. Citation2016), Fragaria virginiana (KY085911), Ficus racemosa (KT368151), Ficus religiosa (KY416513; Bruun-Lund et al. Citation2017), Geum rupestre (NC_037392; Duan et al. Citation2018), Hemiptelea davidii (MK070168; Liu et al. Citation2019), Hippophae rhamnoides (NC_035548; Chen and Zhang Citation2017), Humulus lupulus (KT266264; Vergara et al. Citation2016), Humulus scandens (NC_039730; Zhang et al. Citation2018), Gironniera subaequalis (MH118121; Zhang et al. Citation2018), Lozanella enantiophylla (MH118123; Zhang et al. Citation2018), Malus baccata (KX499859; Zhang et al. Citation2017), Malus domestica (KY818915), Morus alba (KU981119), Morus cathayana (MW465956), Parasponia rugosa (NC_039732; Zhang et al. Citation2018), Potentilla micrantha (HG931056; Ferrarini et al. Citation2013), Prunus dulcis (NC_034696), Pteroceltis tatarinowii (NC_039733; Zhang et al. Citation2018), Rosa maximowicziana (MG727865; Jeon and Kim Citation2019), Rubus crataegifolius (NC_039704; Yang et al. Citation2017), Sorbus ulleungensis (NC_037022), Trema orientalis (NC_039734; Zhang et al. Citation2018), Ulmus davidiana (NC_032718), Ulmus chenmoui (MG581403; Zhang et al. Citation2019), Ziziphus jujuba (NC_030299; Ma et al. Citation2017), Ziziphus mauritiana (NC_037151), and Ziziphus spina-christi (KY628305). Castanea mollissima (KY951992) and Cucurbita moschata (NC_036506) were included as outgroups. The numbers on the nodes indicate bootstrap values from 1000 replicates. The scale bar represents the number of substitutions per site.
Discussion and conclusions
B. racemosa is a rare plant distributed only in a limited region in Jeollabuk-do Province, South Korea. In this study, we reported the complete plastome of B. racemosa together with its genome features. Phylogenetic analysis based on the complete plastome strongly supported earlier study that Rhamnaceae are monophyletic and sister to the Elaeagnaceae family (Cheon et al. Citation2018). Also, B. racemosa is particularly closely related to Berchemia flavescens (BS = 100). This study provides valuable insights into the phylogenetic and evolutionary position of B. racemosa in the Rhamnaceae family and Order Rosales.
Author contributions
Conceptualization, J.P.; methodology, J.P. and J.K.; data analysis, J.K.; investigation, J.P. and J.K.; resources, J.P. and J.K; data curation, J.K.; original draft preparation, J.P. and J.K.; review and editing, J.P.; project administration, J.K.; funding acquisition, J.K. All authors have read and agreed to the published version of the manuscript.
Supplemental Material
Download MS Word (116.6 KB)Disclosure statement
The authors report there are no competing interests to declare.
Data availability statement
The genome sequence data that support the findings of this study are openly available in NCBI (https://www.ncbi.nlm.nih.gov) under the accession no. ON749761. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA835661, SRR21615884, and SAMN30910605, respectively.
Additional information
Funding
References
- Bruun-Lund S, Clement WL, Kjellberg F, Ronsted N. 2017. First plastid phylogenomic study reveals potential cyto-nuclear discordance in the evolutionary history of Ficus L. (Moraceae). Mol Phylogenet Evol. 109:93–104.
- Capella-Gutiérrez S, Silla-Martínez J, Gabaldón T. 2009. TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Chen SY, Zhang XZ. 2017. Characterization of the complete chloroplast genome of seabuckthorn (Hippophae rhamnoides L.). Conservation Genet Resour. 9(4):623–626.
- Cheon KS, Kim KA, Yoo KO. 2018. The complete chloroplast genome sequence of Berchemia berchemiifolia (Rhamnaceae). Mitochondrial DNA B Resour. 3(1):133–134.
- Choi KS, Son O, Park S. 2015. The chloroplast genome of Elaeagnus macrophylla and trnH duplication event in Elaeagnaceae. PLOS One. 10(9):E0138727.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Duan N, Liu S, Liu B. 2018. Complete chloroplast genome of Taihangia rupestris var. rupestris (Rosaceae), a rare cliff flower endemic to China. Conservation Genet Resour. 10(4):809–811.
- Ferrarini M, Moretto M, Ward JA, Šurbanovski N, Stevanović V, Giongo L, Viola R, Cavalieri D, Velasco R, Cestaro A, et al. 2013. An evaluation of the PacBio RS platform for sequencing and de novo assembly of a chloroplast genome. BMC Genomics. 14:670.
- Huang X, Deng T, Lin N, Lv Z, Zhang X, Zhou Z, Wang Y, Sun H. 2019. Complete chloroplast genome sequences of Poikilospermum lanceolatum (Urticeae). Mitochondrial DNA B Resour. 4(2):2131–2132.
- Jeon JH, Kim SC. 2019. Comparative analysis of the complete chloroplast genome sequences of three closely related East-Asian Wild Roses (Rosa sect. Synstylae; Rosaceae). Genes. 10(1):23.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu HB, Li WQ, Xie XM, Zhang XZ, Sun C, Xu JC, Lu YZ. 2019. The complete chloroplast genome of Hemiptelea davidii (Ulmaceae), the only species of the genus Hemiptelea. Mitochondrial DNA Part B. 4(1):229–230.
- Ma Q, Li S, Bi C, Hao Z, Sun C, Ye N. 2017. Complete chloroplast genome sequence of a major economic species, Ziziphus jujuba (Rhamnaceae). Curr Genet. 63(1):117–129.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Oh H, Seo B, Lee S, Ahn DH, Jo E, Park JK, Min GS. 2016. Two complete chloroplast genome sequences of Cannabis sativa varieties. Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2835–2837.
- Shen L, Guan Q, Amin A, Zhu W, Li M, Li X, Zhang L, Tian J. 2016. Complete plastid genome of Eriobotrya japonica (Thunb.) Lindl and comparative analysis in Rosaceae. Springerplus. 5(1):2036.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Vergara D, White KH, Keepers KG, Kane NC. 2016. The complete chloroplast genomes of Cannabis sativa and Humulus lupulus. Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3793–3794.
- Wang YH, Chen SY, Zhang SD. 2018. Characterization of the complete chloroplast genome of Berchemiella wilsonii var. wilsonii (Rhamnaceae), an endangered species endemic to China. Conservation Genet Resour. 10(1):39–41.
- Wu ZY, Du XY, Milne RI, Liu J, Li DZ. 2017. Characterization of the complete chloroplast genome sequence of Cecropia pachystachya. Mitochondrial DNA B Resour. 2(2):735–737.
- Yang JY, Pak JH, Kim SC. 2017. The complete chloroplast genome sequence of Korean raspberry Rubus crataegifolius (Rosaceae). Mitochondrial DNA B Resour. 2(2):793–794.
- Zhang SD, Jin JJ, Chen SY, Chase MW, Soltis DE, Li HT, Yang JB, Li DZ, Yi TS. 2017. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 214(3):1355–1367.
- Zhang H, Jin J, Moore MJ, Yi T, Li D. 2018. Plastome characteristics of Cannabaceae. Plant Divers. 40(3):127–137.
- Zhang X, Wang Y, Wang M, Yuan Q, Huang L. 2020. The complete chloroplast genome of the Crataegus kansuensis (Rosaceae): characterization and phylogeny. Mitochondrial DNA B Resour. 5(3):2920–2921.
- Zhang H, Zhang Q, Fan W, Chen X, Wang J, Li Q, Zhao G. 2019. The complete chloroplast genome of a rare and endangered elm (Ulmus chenmoui) endemic to East China. Conservation Genet Resour. 11(2):169–172.
- Zhao Y, Lu D, Han R, Wang L, Qin P. 2018. The complete chloroplast genome sequence of the shrubby cinquefoil Dasiphora fruticosa (Rosales: Rosaceae). Conservation Genet Resour. 10(4):675–678.
- Zhu XF, Li Y, Lu Z. 2019. The complete chloroplast genome sequence of Berchemia flavescens (Rhamnaceae). Mitochondrial DNA Part B. 4(1):1302–1303.


