Abstract
Belligobio pengxianensis is a small fish endemic to the upper Yangtze River of China. In this study, the complete mitochondrial genome of B. pengxianensis is determined for the first time, and it should become a reference sequence to aid in species identification, biodiversity monitoring and conservation. The mitogenome has overall length of 16,610 bp and AT content of 55.23%, including 13 protein-coding genes, two ribosomal RNAs, 22 transfer RNAs, and one non-coding control region. The results of phylogenetic analyses show that B. pengxianensis is nested within the genus Hemibarbus.
1. Introduction
Fishes of the family Gobionidae (Teleostei: Cypriniformes) include about 28 genera and 226 species widely distributed in drainages across East Asia and Europe (Fricke et al. Citation2022). The gobionid fishes can be divided into three major clades—the Hemibarbus-Squalidus clade, Sarcocheilichthyinae, and Gobioninae—where the Hemibarbus-Squalidus clade is composed of the three genera Belligobio, Hemibarbus, and Squalidus (Tan and Armbruster Citation2018). Belligobio is comprised of three species: B. eristigma Jordan & Hubbs 1925, B. nummifer (Boulenger, 1901), and B. pengxianensis Lu, Luo & Chen 1977 (Bănărescu Citation1992; Yue Citation1998).
B. pengxianensis () is a small (standard length of less than 13 cm) fish (Yue Citation1998; Li Citation2020). In the very few studies (Ding Citation1994; Wang et al. Citation2017), it was considered to be rare, on the basis of its known records only in the Jianjiang River (a tributary of the upper Yangtze River). In a recent study (Li Citation2020), B. pengxianensis has been found to also be distributed in the Fujiang River (a tributary of the upper Yangtze River) based on molecular species delimitation using the mitochondrial cytochrome b gene and eight nuclear loci. Li (Citation2020) has also indicated that B. pengxianensis and B. nummifer could not be differentiated by previously defined diagnostic characteristics (Yue Citation1998), such that prior records on B. nummifer in multiple tributaries of the upper Yangtze River (Ding Citation1994; Lei et al. Citation2015; Chen et al. Citation2017) might be due to misidentification of B. pengxianensis. Therefore, the ambiguities in species identification between B. pengxianensis and B. nummifer, based on morphological characteristics, require molecular evidence to delimit species boundaries.
Figure 1. Female B. pengxianensis from Tongji Town, Pengzhou City, Sichuan Province, China. Photographed by Cuizhang Fu on September 12, 2015.

In this study, the complete mitochondrial genome of B. pengxianensis is determined for the first time. The mitogenome should become a reference sequence to aid in molecular species identification, biodiversity monitoring and conservation, and is also useful in clarifying phylogenetic position of B. pengxianensis.
2. Materials
The specimen of B. pengxianensis was collected in a dead state from Tongji Town, Pengzhou City, Sichuan Province, China (31.1611°N, 103.8394°E) on 12 September 2015. We obtained the sample from the fish catches of a local fisherman. It was placed in the Zoological Museum of Fudan University under the voucher number FDZM-BPPZ20150912-01 (Cuizhang Fu, [email protected]).
3. Methods
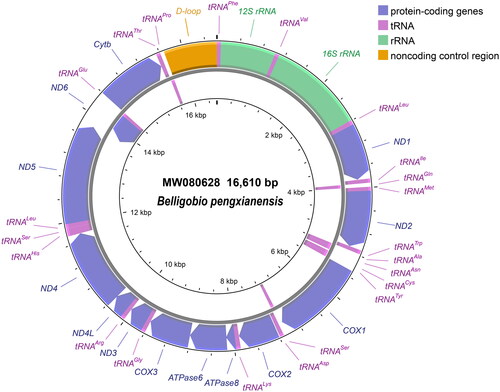
Total genomic DNA was extracted from muscle tissues using a high-salt method (Miller et al. Citation1988). Thirteen primer pairs were used to amplify DNA fragments, with the same PCR system and settings as Chai and Fu (Citation2020). The forward primer of Primer4, reverse primer of Primer6 as well as primers of Primer12 were newly designed in this study (supplemental Table S1 in Appendix I), while the remaining primers have been previously published (Chai and Fu, Citation2020). A total of 13 segments were obtained through Sanger sequencing using the ABI 3730 platform (Applied Biosystems, Foster City, USA). Using a reference genome from B. nummifer (KJ413052; Wang et al. Citation2016), contiguous and overlapping sequences were assembled into mitochondrial genome by Sequencher 5.4 (Gene Codes, Ann Arbor, MI), based on the default parameters. Then, the assembled mitogenome was annotated by MitoAnnotator (Iwasaki et al. Citation2013; http://mitofish.aori.u-tokyo.ac.jp/annotation/input.html). The genome map was drawn using the CGView Server (Grant and Stothard Citation2008).
Table 1. Characteristics of B. pengxianensis mitogenome.
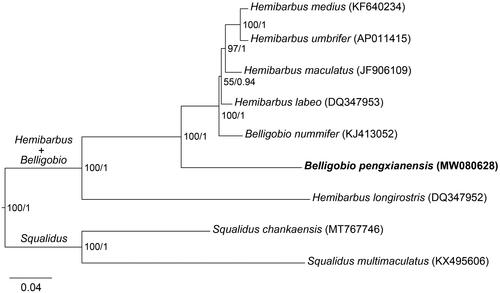
Bănărescu (Citation1992) has treated Belligobio as a sub-genus of Hemibarbus, while Tang et al. (Citation2011) have suggested that Belligobio is a junior synonym of Hemibarbus. Therefore, our phylogenetic analyses included eight mitochondrial genomes from GenBank as follows: B. nummifer (KJ413052; Wang et al. Citation2016), Hemibarbus medius (KF640234; Chen et al. Citation2015), H. umbrifer (AP011415; Tang et al. Citation2011), H. maculatus (JF906109; Li et al. Citation2012), H. labeo (DQ347953; Kim et al. Citation2009), H. longirostris (DQ347952; Kim et al. Citation2009), Squalidus chankaensis (MT767746; Chai and Fu, Citation2020) and S. multimaculatus (KX495606; Yi et al. Citation2017). Our phylogeny was inferred using maximum likelihood (ML) and Bayesian (BI) methods based on 13 protein-coding genes or complete mitochondrial genomes, which the analyses of ML and BI were performed on the software IQ-TREE v2.1.3 (Minh et al. Citation2020) and BEAST v2.6.7 (Bouckaert and Drummond Citation2017), respectively. The ML phylogeny was reconstructed with 1000 ultrafast bootstrap replicates (Hoang et al. Citation2018) and the best models of nucleotide substitution searched by ModelFinder in IQ-TREE. The BI phylogeny was reconstructed with 50,000,000 generations sampled per 1000 generations, and the models of nucleotide substitution were automatically acquired through bModelTest Analyzer in BEAST.
4. Results
A reference Image of B. pengxianensis is provided in . B. pengxianensis is mainly distinguished from other species in the Gobionidae by the combination of three morphological characteristics, as follows: Three row teeth, soft and slim of the last simple dorsal ray, and the scales on the chest hidden under the skin (Yue Citation1998).
The complete mitochondrial genome of B. pengxianensis (MW080628) was assembled from PCR products of 13 segments (supplemental Appendixes I and II). This mitogenome is 16,610 bp in length with the AT content of 55.23%. The circular mitogenome consists of 13 protein-coding genes, two rRNA genes, 22 tRNA genes and one non-coding control region (). The characteristics of the mitogenome are provided in . The protein-coding genes start with ATG/GTG and end with TAA/TAG/TA-/T–. Two conserved blocks of the control region (5′-CAAACCCCCCTACCCCCT-3′ and 5′-TGTCAAACCCCGAAACCA-3′) named CSB-II and CSB-III by Lee et al. (Citation1995), are located in 16,370–16,387 bp and 16,412–16,429 bp, respectively.
The reconstructed phylogenetic trees display consistent topologies under the analyses of ML and BI using 13 protein-coding genes () or complete mitochondrial genomes (supplemental Figure S2 in Appendix III). The results show that B. pengxianensis and B. nummifer don’t form the sister relationships, and all of them are nested within the genus Hemibarbus with high support values.
5. Discussion and conclusion
In this study, we report the mitochondrial genome of B. pengxianensis for the first time. This mitogenome displays the same patterns in the gene compositions, gene arrangements, A + T bias of base compositions, the use of start or stop codon, and conserved sequence blocks of the control region as B. nummifer (Wang et al. Citation2016) and other gobionid species (see, e.g. Kim et al. Citation2009; Li et al. Citation2012; Chai and Fu Citation2020; Tian et al. Citation2022). The phylogenetic nesting of B. pengxianensis and B. nummifer in the genus Hemibarbus was revealed in our phylogeny, bringing into question the validity of the genus Belligobio. In summary, the new mitogenome of B. pengxianensis could contribute to reconstructing inter-specific relationships among species of the genera Hemibarbus and Belligobio, and is also very useful as reference molecular information to support species identification, biodiversity monitoring and conservation in the future.
Ethical approval
Field sampling conformed to the Chinese legislation for scientific purposes. All animal experiments followed Laboratory animal—Guideline for ethical review of animal welfare of the National Standard of the People’s Republic of China (GB/T 35892-2018).
Author contributions
FCZ conceived this study; XB and LMY conducted the experiments; XB and NXM analyzed the data, XB wrote the drafting of the paper; FCZ and NXM revised it critically, and that all authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (1 MB)Supplemental Material
Download MS Word (13.7 MB)Supplemental Material
Download MS Word (402.2 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no.MW080628. Sanger sequencing results are provided in supplementary materials.
Additional information
Funding
References
- Bănărescu PM. 1992. A critical updated checklist of Gobioninae (Pisces, Cyprinidae). Trav Mus Hist Nat. Grigore Antipa. 32:303–330.
- Bouckaert RR, Drummond AJ. 2017. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol Biol. 17(1):42.
- Chai J, Fu CZ. 2020. Three mitochondrial genomes of freshwater fishes in the genus Squalidus (Cypriniformes: Gobionidae). Mitochondrial DNA B Resour. 5(3):3779–3781.
- Chen XWJ, An M, Tang HY, Cao HY, Yang Z. 2017. Length-weight and length-length relationships of seven freshwater fish species from the Daning River, a tributary of the Yangtze River, southwest China. J Appl Ichthyol. 34(3):762–764.
- Chen IS, Liao CR, Shao YT. 2015. The complete mitochondrial genome of Hainanese barbel steed Hemibarbus medius Yue (Teleostei: Cyprinidae). Mitochondrial DNA. 26(5):736–738.
- Ding RH. 1994. The fishes of Sichuan, China. Chengdu City: Sichuan Publishing House of Science and Technology Press.
- Fricke R, Eschmeyer WN, Fong JD. 2022. Eschmeyer’s catalog of fishes: genera/species by family/subfamily. http://researcharchive.calacademy.org/research/ichthyology/catalog/SpeciesByFamily.asp
- Grant JR, Stothard P. 2008. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36(Web Server issue):W181–W184.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Yasunobu M, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. Mitofish and Mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Kim KY, Lim YH, Bang IC, Nam YK. 2009. Phylogenetic relationships among three new Hemibarbus mitogenome sequences belonging to the subfamily Gobioninae (Teleostei, Cypriniformes, and Cyprinidae). Mitochondrial DNA. 20(5–6):119–125.
- Lee WJ, Conroy J, Howell WH, Kocher TD. 1995. Structure and evolution of teleost mitochondrial control regions. J Mol Evol. 41(1):54–66.
- Lei J, Liang YY, Sui XY, Chen YF. 2015. Length-weight relationships for nine fish species of Laohegou River, a tributary of upper Yangtze River, China. J Appl Ichthyol. 31(4):802–803.
- Li P, Xu DD, Peng ZG, Zhang YG. 2012. The complete mitochondrial genome of the spotted steed, Hemibarbus maculatus (Teleostei, Cypriniformes). Mitochondrial DNA. 23(1):34–36.
- Li MY. 2020. Studies on systematics of the family Gobionidae and phylogeny and biogeography of Hemibarbus fishes [PhD dissertation]. Fudan University Shanghai, China.
- Miller SA, Dykes DD, Polesky HF. 1988. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 16(3):1215.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534.
- Tan M, Armbruster JW. 2018. Phylogenetic classification of extant genera of fishes of the order Cypriniformes (Teleostei: Ostariophysi). Zootaxa. 4476(1):6–39.
- Tang KL, Agnew MK, Chen WJ, Hirt MV, Raley ME, Sado T, Schneider LM, Yang L, Bart HL, He SP, et al. 2011. Phylogeny of the gudgeons (Teleostei: Cyprinidae: Gobioninae). Mol Phylogenet Evol. 61(1):103–124.
- Tian W, Ni XM, Fu CZ. 2022. The complete mitochondrial genome of Mesogobio lachneri (Cypriniformes: Gobionidae) from Northeast Asia. Mitochondrial DNA B Resour. 7(10):1810–1813.
- Wang FY, Jean CT, Chen YJ, Lin KY, Liu MY. 2016. The complete mitochondrial genome sequence of Belligobio nummifer (Cypriniformes, Cyprinidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):435–436.
- Wang DG, Shu ZH, Ding RH, Zhou LX. 2017. Endemic Belligobio pengxianensis Lo,Yao & Chen and its survival status. Sichuan J Zool. 36(2):223–226.
- Yi S, Lee SH, Park T, Jang MH. 2017. The complete mitochondrial genome sequence of Squalidus multimaculatus (Cypriniformes: Cyprinidae). Mitochondrial DNA B Resour. 2(2):411–412.
- Yue PQ. 1998. Gobioninae. In: Chen YY, editor. Fauna Sinica, Osteichthyes: Cypriniformes (II). Beijing: Science Press.