Abstract
In the present study, we determined the complete mitochondrial genome of Andreaea regularis Müll. Hal. 1890, a lantern moss of the genus Andreaea Hedw. (Andreaeaceae). The A. regularis mitochondrial genome, with a total length of 118,833 bp, consists of 40 protein-coding genes, 3 ribosomal RNA genes, and 24 transfer RNA genes. A phylogenetic tree constructed with 19 complete mitochondrial genomes composed of liverworts, hornworts, and 15 mosses showed that Andreaeales formed the closest sister to Sphagnales before divergence of the remaining moss groups, indicating A. regularis being one of the earliest mosses. Our findings could be beneficial to investigate the bryophyte evolution.
Introduction
Species of Andreaea Hedw., the type genus of the rock moss order Andreaeales, are frequently referred to as the lantern mosses due to their peculiar appearance, including dehisced sporangia and colors varying from dark-reddish brown to black depending upon the developmental stage (Schofield Citation1985; Ochyra et al. Citation2008). The Andreaea is primarily known as one of the groups of mosses that split the earliest; they are distributed across cold temperate regions but also found in Antarctica (Greene et al. Citation1970; Ochyra et al. Citation2008). To our knowledge, only one mitochondrial genome of the Andreaeales, that of A. wangiana, is available (Huang et al. Citation2019); this lineage split later than another moss lineage, the Sphagnales (Liu et al. Citation2019). Herein, we describe the initial sequencing, annotation, and characterization of the complete mitochondrial genome of A. regularis, contributing to the fundamental genetic knowledge of the genus Andreaea. We also performed a phylogenetic analysis with other closely related moss species to understand the evolutionary relationships of this Antarctic moss species.
Materials and methods
The A. regularis sample was collected in January 2012 by Dr. Hyoungseok Lee from a population growing under natural conditions near the King Sejong Antarctic Station (62° 13′ 26.58″ S; 58° 47′ 05.82″ W) on Barton Peninsula of King George Island. A specimen was deposited at KOPRI Herbarium (https://kvh.kopri.re.kr, Han-Gu Choi, [email protected]) under the voucher number KOPRI-MO00902 and identified as A. regularis by Dr. Young-Jun Yoon. Total genomic DNA extraction and genomic library construction were conducted using a TruSeq DNA Sample Prep Kit (Illumina, San Diego, CA, USA). Whole-genome sequencing was performed using Illumina MiSeq 2 × 300 bp (Illumina) followed by de novo assembly with CLC Genomics Workbench V7.5 (CLC bio, Aarhus, Denmark). All contigs were blasted to the Physcomitrium patens mitochondrial genome (GenBank accession number: NC_007945), and PCR and Sanger sequencing were performed to verify contig ends or ambiguous regions in the contig. To validate the accuracy of the assembly, we mapped trimmed raw sequence reads to the assembled mitochondrial genome to assess the coverage depth (Supplementary Figure 1). Genome annotation was performed using Geneious Prime (version 2022.1.1; Kearse et al. Citation2012), with P. patens (NC_007945) and A. wangiana (NC_046759) as references. The complete mitochondrial genome of A. regularis has been deposited in GenBank under accession number OP067134. A circular mitochondrial map was generated using OGDRAW v1.3.1 (Greiner et al. Citation2019). To evaluate evolutionary relationships, 18 mitochondrial genomes were downloaded from NCBI GenBank: NC_037508 (Bowman et al. Citation2017), NC_043889, NC_024521 (Liu et al. Citation2014), KU725510 (Shaw et al. Citation2016), KU725489 (Shaw et al. Citation2016), NC_046759 (Huang et al. Citation2019), NC_024290 (Bell et al. Citation2014), NC_039775 (Goryunov et al. Citation2021), MK131350, NC_024518 (Liu et al. Citation2014), NC_024514 (Liu et al. Citation2014), NC_007945 (Terasawa et al. Citation2007), NC_026975, NC_027515 (Yoon et al. Citation2016), NC_047462 (Cho et al. Citation2019), NC_027974, NC_012651 (Li et al. Citation2009), NC_037476 (Dong et al. Citation2018). Each bryophyte species used for phylogenetic analysis represents their corresponding family and was selected based on available mitochondrial genome data from GeneBank.
Results and discussion
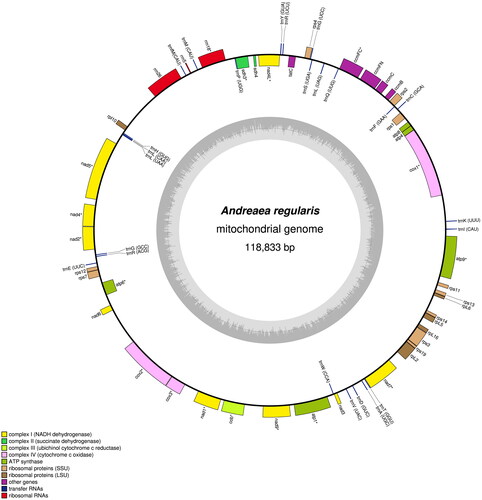
Andreaea regularis is the smallest and most slender of all Antarctic species of the Andreaea genus in Antarctica (Ochyra et al. Citation2008). It differs morphologically from other Andreaea species in terms of its stems, at 5 ∼ 25 mm in length, and ecostate leaves that are lanceolate or ovate-lanceolate, gradually tapering into a narrow long acumen (). The mitochondrial genome of A. regularis has a total length of 118,833 base pairs and consists of 40 protein-coding genes (PCGs), 3 ribosomal RNA genes (rRNAs), and 24 transfer RNA genes (tRNAs) (). There were 10 PCGs (atp1, atp6, ccmFC, cob, cox3, nad2, nad4, nad4L, nad9, sdh3) containing one intron, two PCGs (nad1, nad7) having two introns, three PCGs (atp9, cox2, nad5) having three introns, and one PCG (cox1) having four introns. These cis-splicing genes were verified to be corrected and annotated with multiple sequence alignment (Supplementary Figure 2). The genomic nucleotide composition is 29.4% A, 20.1% C, 21.8% G, and 28.7% T. The start codons for all PCGs comply with ATN rules, except for nad9, rpL16, and tatC; the two former genes employ GTG, and the latter uses ACG. All of the PCGs end with the stop codons TAG, TAA, or TGA, except for ccmFC, which uses CAA. The lengths of the tRNAs range from 71 to 84 bp, totaling 1,799 bp. The lengths of the three rRNAs (rrn5, rrn18, and rrn26) are 122, 1,695, and 3,442 bp, respectively, totaling 5,259 bp. A comparative analysis of the mitochondrial genomes of A. regularis and A. wangiana, which was the only Andreaeaceae mitochondrial genome previously available in GenBank, revealed a significant collinearity for 40 PCGs, 3 rRNAs, and 24 tRNAs. Another unique finding of this analysis was the presence of additional base pairs within three different intergenic regions of A. regularis: (1) 104 bp between trnQ-UUU and trnL-UAG, (2) 111 bp between trnL-UAA and nad5, and (3) 110 bp between nad6 and cox2, partially explaining why the mitogenome size of A. regularis is larger than that of A. wangiana (Supplementary Table 1).
Figure 1. Species reference images of Andreaea regularis; whole plant (a), a shoot (b) and a leaf (c). The pictures were taken by authors in a field on Barton Peninsula of King George Island, Antarctica (a) and in a laboratory (b, c). Scale bar = 1 mm (b) and 0.1 mm (c).

Figure 2. Circular map of the Andreaea regularis mitochondrial genome. Genes outside the outer circle are transcribed clockwise, and those inside the circle are transcribed counterclockwise. Genes belonging to different functional groups are color coded. The innermost darker gray corresponds to GC content, and the lighter gray corresponds to at content.
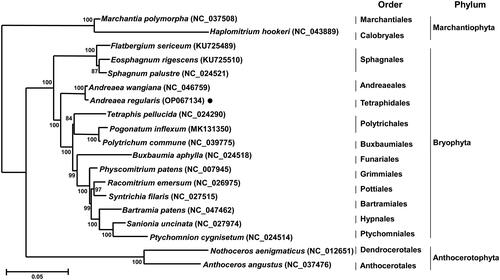
A total of 19 complete mitochondrial genomes, consisting of two hornworts, two liverworts, and 15 mosses (including A. regularis) were aligned using MUSCLE, followed by the construction of a phylogenetic tree based on the maximum-likelihood method (ML; bootstrap repeat is 10,000) using MEGA X software (Kumar et al. Citation2018); the model (GTR + G) with the lowest Bayesian information criterion scores was considered to best specify the substitution pattern (Adachi and Hasegawa Citation1996). As expected, the phylogenetic analysis categorized 19 complete mitochondrial genomes into three different phyla, i.e. Marchantiophyta (liverworts), Anthocerotophyta (hornworts), and Bryophyta (mosses) wherein A. regularis is clustered with A. wangiana within Bryophyta (). More importantly, Andreaeales forms a sister group with Sphagnales (Sphagnum palustre, Eosphagnum rigescens, and Flatbergium sericeum), indicating that Andreaeales is an early diverging lineage in the evolution of mosses. Indeed, Huang et al. (Citation2019) noted that both Sphagnales and Andreaeales represent early diverging mosses based on an evolutionary trend of size reduction of the mitochondrial genome wherein Sphagnales and Andreaeales had relatively higher number of introns and longer intergenic spacers than derived mosses. This notion is further reinforced by our comparative analysis of mitochondrial genome sizes across 15 selected mosses (the same species used for phylogenetic analysis as in ), revealing that the mitochondrial genome size of A. regularis is 21.2 kb smaller than the average size of Sphagnopsida mitochondrial genomes and 13.7 kb larger than the average size of Bryopsida mitochondrial genomes (Supplementary Table 1). The present study is the first report of the complete mitochondrial genome of A. regularis and will be helpful in our understanding of the evolution of mosses, particularly Andreaea.
Ethical approval
The materials used in this study are not included in the IUCN Red List of Threatened Species, and the sampling site is not located in any protected area. The field study was conducted with the permission granted by Minister of Foreign Affairs of the Republic of Korea.
Author contributions
HsL, KM and HdL jointly performed the analysis and wrote the manuscript. YY identified a specimen. PK, YY, and HsL extracted DNA and assembled the mitochondrial genome. SS and KM annotated the mitochondrial genome and carried out the phylogenetic analysis. All authors have reviewed and approved the manuscript.
Supplemental Material
Download Zip (1.1 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. OP067134. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA911323, SRP412618, and SAMN32178634, respectively.
Additional information
Funding
References
- Adachi J, Hasegawa M. 1996. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J Mol Evol. 42(4):459–468. doi: 10.1007/BF02498640.
- Bell NE, Boore JL, Mishler BD, Hyvönen J. 2014. Organellar genomes of the four-toothed moss, Tetraphis pellucida. BMC Genomics. 15(1):383. doi: 10.1186/1471-2164-15-383.
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F, et al. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 171(2):287–304.e15. doi: 10.1016/j.cell.2017.09.030.
- Cho SM, Byun MY, Lee H, Park H, Lee J. 2019. The complete mitogenome of the Antarctic moss Bartramia patens Brid. Mitochondrial DNA B Resour. 4(1):1759–1760. doi: 10.1080/23802359.2019.1610096.
- Dong S, Xue JY, Zhang S, Zhang L, Wu H, Chen Z, Goffinet B, Liu Y. 2018. Complete mitochondrial genome sequence of Anthoceros angustus: conservative evolution of the mitogenomes in hornworts. Bryologist. 121(1):14–22. doi: 10.1639/0007-2745-121.1.014.
- Goryunov DV, Sotnikova EA, Goryunova SV, Kuznetsova OI, Logacheva MD, Milyutina IA, Fedorova AV, Fedosov VE, Troitsky AV. 2021. The mitochondrial genome of nematodontous moss Polytrichum commune and analysis of intergenic repeats distribution among bryophyta. Diversity. 13(2):54. doi: 10.3390/d13020054.
- Greene SW, Greene DW, Browne PD, Pacey PD. 1970. Antarctic moss flora I. The Genera Andreaea, Pohlia, Polytrichum, Psilopilum and Sarconeurum. Sci Rep Br Antarct Surv. 64:1–118.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi: 10.1093/nar/gkz238.
- Huang WZ, Ma WZ, Schneider H, Yu Y, Wu YH. 2019. Mitochondrial genome from Andreaea wangiana reveals structural conservatism and a trend of size reduction in mosses. Bryologist. 122(4):597–606. doi: 10.1639/0007-2745-122.4.597.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi: 10.1093/bioinformatics/bts199.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi: 10.1093/molbev/msy096.
- Li L, Wang B, Liu Y, Qiu YL. 2009. The complete mitochondrial genome sequence of the hornwort Megaceros aenigmaticus shows a mixed mode of conservative yet dynamic evolution in early land plant mitochondrial genomes. J Mol Evol. 68(6):665–678. doi: 10.1007/s00239-009-9240-7.
- Liu Y, Johnson MG, Cox CJ, Medina R, Devos N, Vanderpoorten A, Hedenäs L, Bell NE, Shevock JR, Aguero B, et al. 2019. Resolution of the ordinal phylogeny of mosses using targeted exons from organellar and nuclear genomes. Nat Commun. 10(1):1485. doi: 10.1038/s41467-019-09454-w.
- Liu Y, Medina R, Goffinet B. 2014. 350 My of mitochondrial genome stasis in mosses, an early land plant lineage. Mol Biol Evol. 31(10):2586–2591. doi: 10.1093/molbev/msu199.
- Ochyra R, Lewis Smith RI, Bednarek-Ochyra H. 2008. The illustrated moss flora of Antarctica. New York (NY): Cambridge University Press; p. 73–87.
- Schofield WB. 1985. Introduction to bryology. New York (NY): Macmillan.
- Shaw AJ, Devos N, Liu Y, Cox CJ, Goffinet B, Flatberg KI, Shaw B. 2016. Organellar phylogenomics of an emerging model system: sphagnum (peatmoss). Ann Bot. 118(2):185–196. doi: 10.1093/aob/mcw086.
- Terasawa K, Odahara M, Kabeya Y, Kikugawa T, Sekine Y, Fujiwara M, Sato N. 2007. The mitochondrial genome of the moss Physcomitrella patens sheds new light on mitochondrial evolution in land plants. Mol Biol Evol. 24(3):699–709. doi: 10.1093/molbev/msl198.
- Yoon YJ, Kang Y, Kim MK, Lee J, Park H, Kim JH, Lee H. 2016. The complete mitochondrial genome of an Antarctic moss Syntrichia filaris (Müll. Hal.). Mitochondrial DNA A DNA Mapp. Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2779–2780. doi: 10.3109/19401736.2015.1053062.