Abstract
Macropodus hongkongensis (Freyhof & Herder, 2002), is sparsely distributed in Hong Kong and Guangdong provinces. Recently, a new geographical population of M. hongkongensis was discovered in the Wanquan River in the Hainan province. Therefore, this study focused on sequencing the complete mitochondrial genome of the new geographical population of Macropodus hongkongensis from the Wanquan River. The circular mtDNA molecule was 16,492 bp in size, and the overall base compositions were A (30.30%), C (24.90%), T (29.80%), and G (15.00%), with a slight bias toward A + T. The complete mitogenome encoded 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a control region. Phylogenetic analysis indicated that M. hongkongensis of the Hainan Wanquan River was most closely related to the M. hongkongensis of the Gongdong population. These results provide useful genetic information for species identification and phylogenetic studies of Macropodus species.
Introduction
Macropodus hongkongensis, commonly known as the Hong Kong paradise fish, belonging to the genus Macropodus of the family Belontiida and is sparsely distributed in Hong Kong and Guangdong provinces (Chan et al. Citation2008; Wang et al. Citation2009). Recently, a new geographical population of M. hongkongensis was discovered in the Wanquan River in the Hainan province. The known geographical distribution area of M. hongkongensis in Hainan is limited, and its habitat consists of small streams in the intermountain thickets (Shen et al. Citation2021). In this study, the complete mitogenome of M. hongkongensis of the Wanquan River, a new geographical location, was determined and its phylogenetic relationship was analyzed.
Materials and methods
Specimens of adult M. hongkongensis () were collected from the Wanquan River, Hainan province (19°11′49.36″N, 110°47′60.59 E) and deposited in the Hainan Academy of Ocean and Fisheries (http://www.hnhky.cn/, Qingfeng Zhang, [email protected]) under the voucher number HNFF0930101. Total DNA was extracted using an Ezup DNA kit and sequenced using Illumina 6000. The genome was assembled using the GetOrganelle (Jin et al. Citation2020). The annotated sequence was submitted to GenBank with the accession number OQ630878.1, and the read coverage depth map is shown in Figure S1 (Supplementary material). A phylogenetic tree was constructed by software RAxML (Stamatakis Citation2014), using sequences from Genbank: M. hongkongensis (MN128300.1), M. erythropterus (NC_029946.1), M. opercularis (NC_025932.1), Betta apollon (LC433688.1), Trichopodus pectoralis (KY606170.1), Trichogaster trichopterus (KP100265.1), Colisa fasciata (KP301136.1), Colisa lalia (AP006039.1), (Liu et al. Citation2019); Betta splendens (AB571120.1), (Yu et al. Citation2016); Epinephelus hexagonatus (NC_065829.1), (Wang et al. Citation2022); Alces alces (JN632595.1), (Hassanin et al. Citation2012); Centropyge eibli (KT001113.1) (Shen et al. Citation2016).
Results
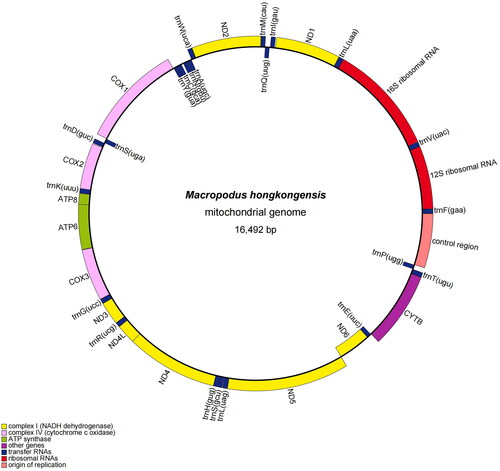
The complete circular mitochondrial genome of M. hongkongensis of the new population was 16,492 bp in length () with a base composition of 30.30, 24.90, 29.80, and 15.00% of A, C, T, and G, respectively. The composition showed a slight bias toward A + T content (60.10%), which was consistent with other Macropodus fishes (Xu et al. Citation2016). The mitogenome contained 13, 22, and 2; protein (PCGs), tRNA and rRNA-coding genes, respectively, along with a control region. All PCGs had an ATG start codon, except COX1 (GTG) and ND6 (CTA). Four PCGs (ND1, ATP6, ATP8, and ND5) terminated with TAA, and eight PCGs (ND2, COX2, COX3, ND3, ND4L, ND4, ND6, and CYTB) ended with the incomplete stop codon TA– or T––. Except for the ND6 and the eight tRNA genes, all other genes were encoded on the heavy strand (H).
Conclusion
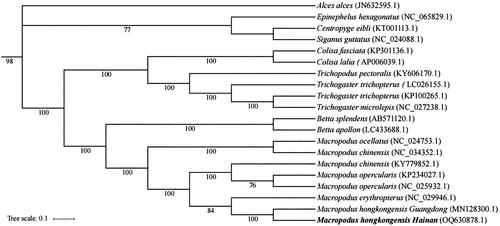
In the phylogenetic analysis, M. hongkongensis of the Hainan population was most closely related to the Guangdong population () and the nucleotide sequence divergence of 13 protein coding genes between the two population was 4.0%, smaller than the interspecific distance of Macropodus erythropterus (13.9%). These results offer molecular markers and a reference for conservation research of M. hongkongensis in different populations.
Author contributions
Zhixin Shen designed the study and planned the experiments. Qingfeng Zhang drafted the manuscript and performed the data analysis. Fangyuan Li and Gaojun Li conceptualized and revised the manuscript. Yang Dong, Ji Wang, and Dikai Chen were responsible for sample collection, data acquisition, and revision of the manuscript. All authors agree to be accountable for all aspects of this study.
Ethical approval
This study did not require ethical approval or permission to collect samples.
Supplemental Material
Download TIFF Image (3.1 MB)Acknowledgement
We would like to thank Editage (www.editage.cn) for English language editing.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank at [https://www.ncbi.nlm.nih.gov] under the accession number: OQ630878.1. The associated **BioProject**, **SRA**, and **Bio-Sample** numbers are PRJNA955001, SRR24305076, and SAMN34163124, respectively.
Additional information
Funding
References
- Chan BPL, Dudgeon D, Chen XL. 2008. Threatened fishes of the world: Macropodus hongkongensis Freyhof and Herder, 2002 (Osphronemidae). Environ Biol Fish. 81(4):367–368. doi:10.1007/s10641-007-9210-0.
- Hassanin A, Delsuc F, Ropiquet A, Hammer C, Jansen van Vuuren B, Matthee C, Ruiz-Garcia M, Catzeflis F, Areskoug V, Nguyen TT, et al. 2012. Pattern and timing of diversification of Cetartiodactyla (Mammalia, Laurasiatheria), as revealed by a comprehensive analysis of mitochondrial genomes. C R Biol. 335(1):32–50. doi:10.1016/j.crvi.2011.11.002.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Liu H, Xu N, Yang J, Zhang Q, Zuo K, Lv F. 2019. The complete mitogenome of Hong Kong paradise fish (Macropodus hongkongensis), an endemic freshwater fish in South China. Mitochondrial DNA B Resour. 4(2):2849–2850. doi:10.1080/23802359.2019.1660271.
- Shen KN, Chang CW, Chen CH, Hsiao CD. 2016. Complete mitogenomes of multicolor angelfish (Centropyge multicolor) and Yellowhead angelfish (Centropyge joculator) (Teleostei: Pomacanthidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2807–2808. doi:10.3109/19401736.2015.1053077.
- Shen ZX, Wang DQ, Li GJ, Cai XW, Hu YC, Zhu H, Tong YN, Gu DE, LiF Y, Zhao GJ, et al. 2021. Illustrated hand-book of freshwater and estuarine fishes in Hainan. Beijing (China): China Agriculture Press; p. 232.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi:10.1093/bioinformatics/btu033.
- Wang C, Ye P, Liu M, Zhang Y, Feng H, Liu J, Zhou H, Wang J, Chen X. 2022. Comparative analysis of four complete mitochondrial genomes of Epinephelidae (Perciformes). Genes (Basel). 13(4):660. doi:10.3390/genes13040660.
- Wang PX, Luo JR, Bai JJ, Ye X, Hu Y-C, Wang X-J. 2009. Cytochrome b complete sequences and RAPD analysis of the phylogenetic relationships among the fishes of the genus Macropodus. Chin J Zool. 44(5):14–23.
- Xu HX, Ma ZH, Yang RB, Xie LP, Yang XF. 2016. Complete mitochondrial genome of Macropodus ocellatus (Perciformes: Anabantidae: Macropodusinae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1105–1106. doi:10.3109/19401736.2014.933321.
- Yu P, Ding S, Yang Q, Bi Z, Chen L, Liu X, Song X, Wan Q. 2016. Complete sequence and characterization of the paradise fish Macropodus erythropterus (Perciformes: Macropodusinae) mitochondrial genome. Mitochondrial DNA B Resour. 1(1):54–55. doi:10.1080/23802359.2015.1137820.