?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The real-time requirement for image segmentation in laparoscopic surgical assistance systems is extremely high. Although traditional deep learning models can ensure high segmentation accuracy, they suffer from a large computational burden. In the practical setting of most hospitals, where powerful computing resources are lacking, these models cannot meet the real-time computational demands. We propose a novel network SwinD-Net based on Skip connections, incorporating Depthwise separable convolutions and Swin Transformer Blocks. To reduce computational overhead, we eliminate the skip connection in the first layer and reduce the number of channels in shallow feature maps. Additionally, we introduce Swin Transformer Blocks, which have a larger computational and parameter footprint, to extract global information and capture high-level semantic features. Through these modifications, our network achieves desirable performance while maintaining a lightweight design. We conduct experiments on the CholecSeg8k dataset to validate the effectiveness of our approach. Compared to other models, our approach achieves high accuracy while significantly reducing computational and parameter overhead. Specifically, our model requires only 98.82 M floating-point operations (FLOPs) and 0.52 M parameters, with an inference time of 47.49 ms per image on a CPU. Compared to the recently proposed lightweight segmentation network UNeXt, our model not only outperforms it in terms of the Dice metric but also has only 1/3 of the parameters and 1/22 of the FLOPs. In addition, our model achieves a 2.4 times faster inference speed than UNeXt, demonstrating comprehensive improvements in both accuracy and speed. Our model effectively reduces parameter count and computational complexity, improving the inference speed while maintaining comparable accuracy. The source code will be available at https://github.com/ouyangshuiming/SwinDNet.
Introduction
Laparoscopic surgery is a minimally invasive surgical technique that has been widely adopted. Compared to traditional surgery, laparoscopic surgery offers advantages such as smaller incisions, minimal damage to surrounding tissues, faster postoperative recovery, and reduced pain. Laparoscopic surgery has gained widespread popularity. The laparoscopic camera captures images, which are transmitted to a signal processing system via fiber optics using digital imaging technology. These real-time video images are displayed on a dedicated monitor. The surgeon then performs the surgery using specialized laparoscopic instruments based on the visual information displayed on the monitor screen.
Clearly, computer assistance and surgical navigation in laparoscopic surgery require accurate segmentation of relevant organs to generate organ mask for stereo surface reconstruction for intraoperative to preoperative organ surface registration, which can be achieved through semantic segmentation techniques. Semantic segmentation is a pixel-level dense segmentation method that classifies each pixel in an image into target or background classes. It is a classical and fundamental problem in the field of computer vision. Semantic segmentation has a wide range of applications and has been successfully applied in various fields such as autonomous driving, remote sensing systems, and medical image diagnosis. Medical image semantic segmentation poses unique complexities and challenges. For instance, obtaining annotated datasets can be difficult. Optical imaging is often affected by irregular factors such as occlusion, shadows, uneven lighting, and noise, making segmentation of medical images more challenging compared to natural images.
In recent years, the rapid development of deep learning has greatly improved the performance of semantic segmentation, and several deep learning-based methods have shown promising results in medical image segmentation tasks. In 2015, Rosenberg et al. proposed U-Net [Citation1], which adopts an encoder-decoder architecture with skip connections. U-Net has become a benchmark model for many medical image segmentation tasks. U-Net++ [Citation2] replaces the long skip connections in U-Net with short skip connections to better integrate features at different scales and reduce the scale discrepancy between feature maps. ResU-Net [Citation3] introduces residual connections into U-Net, which improves gradient flow in the network, prevents network degradation, and accelerates network convergence. Attention U-Net [Citation4] incorporates attention gates into U-Net, which utilize attention modules to adjust the output features of the encoder before concatenating them with corresponding features in the decoder. This helps suppress irrelevant regions and highlight salient features in specific local areas. Additionally, there is DeepLabv3 + [Citation5], which combines an encoder-decoder structure with atrous convolution and ASPP (Atrous Spatial Pyramid Pooling) to optimize edge accuracy. It also achieves a fusion of encoder-decoder and multi-scale networks. DeepLabv3+ has been a representative model at the state-of-the-art (SOTA) level in semantic segmentation for a long time. TransUNet [Citation6] combines the popular Transformer architecture with U-Net. It uses Transformers to encode feature maps into input sequences, extracting global contextual information. The decoder performs upsampling on the encoded features and combines them with high-resolution feature maps for precise localization.
There have been several relevant works on semantic segmentation of laparoscopic images in recent years. Kolbinger et al. [Citation7] conducted experiments on the Dresden Surgical Anatomy Dataset using the DeepLabv3 model and SegFormer [Citation8] model. They found that the DeepLabv3 model needs improvement in terms of accuracy, while the real-time performance of the SegFormer model is not satisfactory. In [Citation9], the authors discussed how to perform laparoscopic image segmentation in a semi-supervised manner when there is insufficient annotated data. They achieved significant segmentation accuracy without using additional labeled data but instead utilizing more unlabeled data. In [Citation10], the authors proposed a deep residual network for automatic liver segmentation in laparoscopic images. This network is a fully convolutional neural network (CNN) and demonstrated that CNNs can accurately segment the liver and other anatomical structures in laparoscopic images. The paper [Citation11] explores the performance of different neural networks, loss functions, and training strategies for semantic segmentation of various organs and tissues in laparoscopic images. The neural networks considered include TernausNet [Citation12], SegNet [Citation13], LinkNet [Citation14], among others, while the loss functions include Soft-Jaccard, generalized Dice, and cross-entropy (CE) losses.
The aforementioned methods, while achieving good segmentation accuracy, suffer from large parameter sizes, computational complexity, and long inference time. They also have high requirements for computational power, which is often lacking in hospitals compared to research laboratories. Consequently, these models are not suitable for real-time requirements in laparoscopic surgery. To reduce the complexity and size of models while maintaining network accuracy, lightweight networks have been designed by appropriately reducing the number of channels and weight parameters in convolutional layers. This reduction aims to decrease the floating-point computations, enhance the model’s prediction speed, and improve efficiency. Lin et al. proposed to replace the fully connected layers with a global average pooling (GAP) layer in the Network-in-Network architecture [Citation15]. This clever design effectively reduces the original parameter count of the network. Chollet [Citation16] introduced Xception, which combines deep convolutions with 1 × 1 pointwise convolution to extract channel and spatial information separately. In the same year, Iandola et al. presented a lightweight network model called SqueezeNet [Citation17], which compresses the network by reducing the size of convolutional kernels and the number of channels. Howard et al. proposed MobileNet [Citation18] based on the concept of depthwise separable convolution, which separates a standard convolution into depthwise convolutions and pointwise convolutions. Zhang et al. introduced ShuffleNet [Citation19], by introducing the concept of group convolution, which groups channels and computes them with different convolutional kernels to reduce the computational cost of convolutions. Huang et al. proposed DenseNet, which directly connects layers to capture more inter-layer information and incorporates learnable group convolutions that undergo sparse operations during training, significantly reducing the number of parameters and computations. Sandler et al. improved MobileNet with the introduction of MobileNetV2 [Citation20], which introduced linear bottleneck units and inverted residual blocks. Ma et al. conducted a comparative study of ShuffleNetv1 and MobileNetV2, considering factors such as FLOPs, memory usage, and model parallelism, and proposed four guidelines for efficient network architecture design. Based on these guidelines, they improved ShuffleNetv1 and proposed ShuffleNetv2 [Citation21], drawing inspiration from DenseNet. Google applied neural architecture search techniques to design MobileNetv3 [Citation22] by applying average pooling and removing the final convolutional layer, along with the use of the h-swish activation function. In the same year, Tan et al. employed neural architecture search techniques to develop EfficientNets [Citation23]. Valanarasu et al. proposed UNeXt, where a tokenized MLP block with shift-based MLPs significantly reduces complexity and parameter count [Citation24].
In this work, our focus is on designing a network that maintains good segmentation performance while being more efficient, with fewer parameters and faster inference time. Ultimately, we propose a novel network called SwinD-Net, which incorporates Skip connections, Depth-wise separable convolutions, and Swin Transformer Blocks. We evaluated our model on the CholecSeg8k [Citation25] dataset and compared it with U-Net, DeepLabv3+, U-Net++, Attention U-Net, MobileNetV3, and UNeXt [Citation26].
In this study, the following three main contributions were made: (1) We propose a novel and fast segmentation network model that combines deep convolution and Swin Transformer Blocks. (2) In feature extraction, we adopt deep convolutions to enhance speed and introduce Swin Transformer Blocks to capture global information and high-level semantic features, thus maintaining segmentation accuracy. (3) Compared to non-lightweight models, we successfully reduce computational complexity and the number of parameters while maintaining almost the same level of accuracy. This significantly speeds up the inference process.
Method and materials
Network architecture
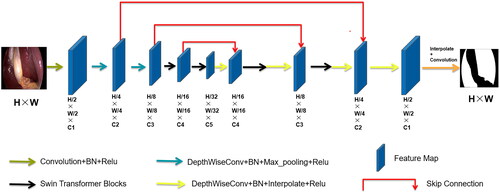
Based on the encoder-decoder structure and skip connections of U-Net, we replaced some convolution operations with depthwise separable convolutions and introduced Swin Transformer Blocks to design our lightweight segmentation network, SwinD-Net. Additionally, we discovered through experiments that removing the skip connection from the first layer does not significantly affect the accuracy, indicating that shallow texture features have minimal impact on the final segmentation results for large object segmentation tasks. Furthermore, we found that reducing the number of channels in the shallow feature maps has minimal impact on accuracy for this task, while significantly reducing the number of parameters. Reducing the resolution of the input image is an important approach to improving the speed of image segmentation. However, directly downsampling the image using interpolation results in information loss and decreases the segmentation accuracy. To address this problem, we employed linear mapping to transform the image data into a high-order feature representation. Specifically, we used a convolutional layer with a kernel size of 3 × 3 and a stride of 4 to implement this linear mapping. This approach preserves image information without compromising segmentation accuracy and achieves a reduced image resolution, thereby speeding up the segmentation process. To improve the accuracy of the network, we replaced convolution operations with depthwise separable convolutions. Considering that liver segmentation involves a large object, a larger receptive field implies higher accuracy. Therefore, we introduced Swin Transformer Blocks to obtain a global receptive field and extract global features, thus enhancing liver segmentation accuracy.
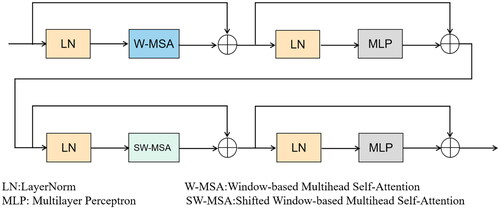
Overall, in the encoder stage, after extracting features with a regular convolutional layer, the obtained feature map undergoes two depthwise separable convolutions for further feature extraction. Depthwise Separable Convolution (DSC) consists of two parts: Depthwise (DW) convolution and Pointwise (PW) convolution. In depthwise convolution, each channel is convolved with a separate kernel, where each channel is convolved by only one kernel. Since the number of output channels in the convolution operation is equal to the number of kernels, N feature maps with a channel size of 1 are obtained. These output feature maps from all kernels are then concatenated to produce an output feature map with N channels. Pointwise convolution, in essence, is a 1 × 1 convolution that allows for freely changing the number of output feature map channels and performs channel fusion on the feature maps obtained from the main channel convolution. After the two depthwise separable convolutions, the feature map is passed through Swin Transformer Blocks. As shown in , Swin Transformer Blocks consist of two consecutive Transformer modules. The first module utilizes a Window-based Multihead Self-Attention (W-MSA) mechanism, while the second module employs a Shifted Window-based Multihead Self-Attention (SW-MSA) mechanism. Information exchange between windows is achieved through a shifted window mechanism. W-MSA (Window-based Multihead Self-Attention) confines attention within a window, reducing computational complexity and memory consumption, and is capable of capturing local features in an image. Meanwhile, in SW-MSA (Shifted Window-based Multihead Self-Attention), by shifting the windows, it can capture the relationships between windows, enabling the acquisition of long-range dependencies.
The computational complexity of MSA and W-MSA can be approximately estimated as follows:
(1)
(1)
(2)
(2)
h represents the height of the feature map, w represents the width of the feature map, C represents the number of channels in the feature map, M represents the size of each window or patch used in the attention mechanism.
After passing through two Swin Transformer Blocks, the feature map enters the decoder stage. At this stage, a depthwise separable convolution is applied first, followed by a sequence of two combined operations: Swin Transformer Blocks and depthwise separable convolutions. Finally, another depthwise separable convolution is performed, followed by upsampling to obtain the output feature map. The overall network architecture diagram is illustrated below :
Loss function and optimizer
We employed the Adam optimizer, which combines the advantages of the AdaGrad and RMSProp optimization algorithms. Adam optimizer considers both the first and second moments of the gradients to calculate the update step size. This enables it to adapt to sparse gradients and alleviate gradient oscillation.
The loss function we used is the cross-entropy loss function, which is formulated as follows:
(3)
(3)
Dataset
The dataset used in our experiments is the CholecSeg8k dataset, which is a semantic segmentation dataset based on the Cholec80 dataset [Citation27] for laparoscopic cholecystectomy. The Cholec80 dataset consists of 80 videos of cholecystectomy procedures. Annotations for 13 semantic classes are provided at the pixel level for each image, with a resolution of 854 × 480. The provider of the CholecSeg8k dataset selected 17 videos, resulting in a total of 8080 images and masks.
Implementation details
Based on Python 3.7 and PyTorch 1.8, our experiments were conducted on Ubuntu Linux 20.04 OS with an NVIDIA RTX 3090 GPU of 24GB memory. We first do a five-fold cross-validation, four portions (or 80%) serve as the training and validation sets, with one portion (or 20%) being the testing set. And for the final model construction, we divided the dataset into training, validation, and test sets in a 7:1:2 ratio, the training set accounts for 70%, containing 5656 images; the validation set accounts for 10%, containing 808 images; and the testing set accounts for 20%, containing 1616 images.
Results
Evaluation metrics
IoU: Intersection over Union, also known as the Jaccard Index, is calculated as the intersection of the predicted area and the real area divided by the union of the predicted area and the real area.
Dice coefficient: It is used to measure the similarity between two sets and its values range from 0 to 1. Given X and Y as two sets, the Dice coefficient is calculated as follows:
(4)
(4)
FLOPs: Floating Point OPerations, which refers to the number of floating-point arithmetic operations performed by a model. It is used to measure the computational complexity of the model.
Params: The total number of trainable parameters in the network model. It represents the number of learnable weights and biases that need to be updated during the training process.
Inference Time: The time required for the model to process input data and generate output results. It takes into account the model’s complexity, hardware device performance, and optimization strategies. Accurate measurement of inference time allows for a better evaluation of the model’s speed performance in real-world scenarios, helping determine whether it meets real-time requirements or resource constraints.
Experiment results
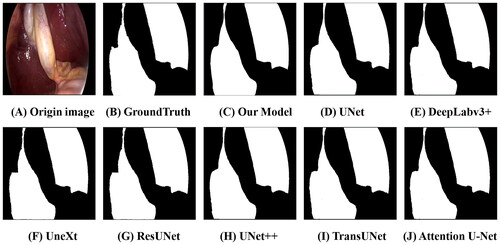
We compared our SwinD-Net model with several classical network models in terms of IoU, Dice score, FLOPs, and Params. The comparison includes state-of-the-art networks such as Deeplabv3+, various variations of U-Net and its family, TransUNet based on Transformer, and the fast segmentation network MobileNetV3, UNeXt. We first listed our 5-fold cross validation experiment result in the Dice coefficient in . We summarized the final results in , and it can be observed that our SwinD-Net model surpasses the lightweight and fast segmentation network UNeXt in terms of Dice score, while having less than 1/20 of the FLOPs and only 1/3 of the parameters compared to UNeXt. Additionally, we found that lightweight networks tend to have slightly lower Dice scores compared to non-lightweight networks, but the difference is negligible, as shown in the comparison of prediction results in , where no statistically significant differences are observed, we conducted paired t-tests comparing the Dice scores of our model with those of other models, and found that all p-values were greater than .05, indicating no significant differences. However, compared to non-lightweight models, our lightweight model has significant advantages in terms of FLOPs and parameters. Our FLOPs are only 1/2000 of U-Net, and the number of parameters is only 1/64 of U-Net. Compared to TransUNet, our model has FLOPs that are only 1/1200 of TransUNet and a parameter count that is only 1/186. Compared to the lightweight segmentation network UNeXt, our model outperforms in all four aspects, with a Dice score lead over UNeXt and parameter count only 1/3 of UNeXt, while the FLOPs are only 1/22 of UNeXt.
Table 1. The five cross-validation comparisons result in a dice coefficient.
Table 2. The segmentation performance in dice coefficient, FLOPs and params of our model and other SOTA model comparison. All p-values of paired tests were greater than 0.05.
Although the number of parameters and FLOPs can be used to assess the computational complexity of a model to some extent, the real indicator that reflects the model’s speed performance in practical applications is the inference time. The number of parameters and FLOPs primarily measure the model’s scale and computational requirements, but they do not directly consider factors such as the actual running speed of hardware devices, data transmission, and delays in the computation process. Therefore, in order to comprehensively evaluate the model’s performance, the inference time must also be considered as a metric. For lightweight models in particular, the calculation of inference time is crucial because these models are typically designed to run in resource-constrained environments such as mobile devices or embedded systems.
When calculating the inference time, we adopted the following timing strategy to minimize the impact of time delay caused by calling time functions: We read in an image, record the start time, then loop the inference of this image ten thousand times, and record the end time after the loop. By looping the inference ten thousand times with only two calls to the time function, we can obtain the actual time spent on inference to the greatest extent. Since most hospital computing devices do not have GPUs, we not only calculated the average inference time on the server GPU but also on the CPU. This allows us to simulate real surgical scenarios in hospitals to the maximum extent. Using an Intel(R) Xeon(R) W-2245 processor with a base speed of 3.90 GHz, we performed inference on 512 × 512 images, and the average inference times for each model are in :
Table 3. The inference time comparison of our model and other SOTA methods on GPU and CPU.
Clearly, our model has the shortest inference time. On the CPU, our average inference time is only 47.49 milliseconds, which is 1/40 of U-Net, 1/14 of DeepLabv3+, 3.5 times faster than MobileNetV3, and 2.4 times faster than UNeXt. Such inference speed is sufficient to meet real-time inference requirements.
Ablation study
Shallow layers primarily extract low-level semantic information. However, for the segmentation of large objects like the liver, deep layers containing high-level semantic information are more important. The Swin Transformer Blocks have the ability to extract global information and effectively capture the relationships between local features, which greatly impact the segmentation accuracy. In this section, we explored the impact of depthwise separable convolutions and Swin Transformer Blocks on the segmentation accuracy of our model in . After the initial feature extraction using deep convolutions in our proposed model, the feature maps are further processed by the Swin Transformer Blocks to extract global information. When replacing depthwise separable convolutions with regular convolutions, we observed that the accuracy almost remained unchanged, but this substitution led to a significant increase in the number of parameters. Regarding the Swin Transformer Blocks, a structure used for extracting global features, it is crucial for tasks like segmenting large objects such as the liver, where global information is vital. Removing this module resulted in a noticeable drop in accuracy, thereby proving that using depthwise separable convolutions instead of regular convolutions, and incorporating Swin Transformer Blocks into our model, are critical steps.
Table 4. The ablation experiment result of our model.
Generalizability experiment
To verify the generalization ability of our model, we directly tested the model well trained on the CholecSeg8K dataset on the Dresden surgical anatomy dataset (DSAD) [Citation28], which has a higher level of segmentation difficulty. Some failed segmentation cases shown in :
Figure 4. Some segmentation failure cases in the DSAD dataset. The ground truth segmentation is colored in green, and our model’s segmentation results are colored in red.
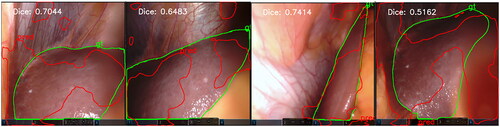
The Dresden dataset is much more challenging to segment than the CholecSeg8K dataset, the average liver segmentation Dice accuracy of state-of-the-art DeepLabv3 and SegFormer model are 0.80 ± 0.23 and 0.83 ± 0.21, respectively [Citation7]. This indicates that our model’s generalization capabilities are not yet excellent, which is a common issue with many convolutional neural network models, such as the decline in the segmentation performance for patient data that is not in the training set [Citation29]. Therefore, our next step is to work on improving the generalization capabilities of our model, such as by using larger models [Citation30,Citation31] to improve generalizability, or employing domain adaptation with model knowledge distillation and small zero-shot learning [Citation32]. This will enable our model to maintain excellent segmentation performance across different datasets, especially those with higher segmentation difficulty.
Conclusion
In this study, we have designed a novel network called SwinD-Net. By incorporating skip connections, deep convolution, and Swin Transformer Blocks, the network achieves high segmentation accuracy while significantly reducing the computational complexity and improving inference speed. Compared to networks such as U-Net and TransUNet, SwinD-Net greatly reduces the floating-point operations and parameter count, and outperforms the lightweight segmentation network UNeXt in multiple evaluation metrics. SwinD-Net utilizes deep convolution in the shallow layers to replace traditional convolution, reducing the number of channels and omitting the first skip connection layer, thereby saving computational and parameter costs. The subsequent introduction of Swin Transformer Blocks captures global information and high-level semantic features, ensuring segmentation accuracy. Our model achieves high accuracy with minimal computational complexity and parameter count, while meeting the requirements for real-time inference speed. It has excellent prospects for applications in scenarios where most hospitals lack powerful computational resources, ensuring real-time segmentation with high accuracy.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation In International Conference on Medical Image Computing and Computer-Assisted Intervention, LNCS vol.9351. 2015. pp. 1–9.
- Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, et al. UNet++: a nested U-net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support, LNCS. vol. 11045; 2018. pp. 3–11.
- Diakogiannis FI, Waldner F, Caccetta P, et al. ResUNet-a: a deep learning framework for semantic segmentation of remotely sensed data. ISPRS J Photogramm. 2020;162:94–114. doi: 10.1016/j.isprsjprs.2020.01.013.
- Oktay O, Schlemper J, Folgoc LL, et al. Attention U-Net: learning where to look for the pancreas. arXiv preprint. arXiv:1804.03999. 2018.
- Chen LC, Zhu Y, Papandreou G, et al. Encoder-decoder with atrous separable convolution for semantic image segmentation In Proceedings of the European Conference on Computer Vision (ECCV). 2018. pp. 801–818.
- Chen J, Lu Y, Yu Q, et al. TransUNet: transformers make strong encoders for medical image segmentation. arXiv Preprint. 2021.
- Kolbinger FR, Rinner FM, Jenke AC, et al. Anatomy segmentation in laparoscopic surgery: comparison of machine learning and human expertise-an experimental study. Int J Surg. 2023;109(10):2962–2974. doi: 10.1097/JS9.0000000000000595.
- Xie E, Wang W, Yu Z, et al. SegFormer: simple and efficient design for semantic segmentation with transformers. Adv Neural Inf Process Syst. 2021;34:12077–12090.
- Fu Y, Robu MR, Koo B, et al. More unlabelled data or label more data? A study on semi-supervised laparoscopic image segmentation. Domain Adaptation and Representation Transfer and Medical Image Learning with Less Labels and Imperfect Data: First MICCAI Workshop, DART 2019, and First International Workshop, MIL3ID 2019, Shenzhen, Held in Conjunction with MICCAI 2019, Shenzhen, China, October 13 and 17, 2019, Proceedings 1. Springer International Publishing. Springer; 2019. pp. 173–180.
- Gibson E, Robu MR, Thompson S, et al. Deep residual networks for automatic segmentation of laparoscopic videos of the liver. In SPIE Medical Imaging 2017: Image-Guided Procedures, Robotic Interventions, and Modeling. vol. 10135. 2017. pp 423–428. doi: 10.1117/12.2255975.
- Scheikl PM, Laschewski S, Kisilenko A, et al. Deep learning for semantic segmentation of organs and tissues in laparoscopic surgery. Curr Dir Biomed Eng. 2020;6(1):20200016. doi: 10.1515/cdbme-2020-0016.
- Iglovikov V, Shvets A. Ternausnet: u -net with vgg11 encoder pre-trained on imagenet for image segmentation. arXiv preprint. arXiv:1801.05746. 2018.
- Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(12):2481–2495. doi: 10.1109/TPAMI.2016.2644615.
- Chaurasia A, LinkNet CE. Exploiting encoder representations for efficient semantic segmentation. In: IEEE Visual Communications and Image Processing (VCIP). IEEE; 2017.
- Lin M, Chen Q, Yan S. Network in network. arXiv:1312.4400. 2013.
- Chollet F. Xception: Deep learning with depthwise separable convolutions In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition (CVPR). 2017. pp. 1251–1258.
- Iandola FN, Han S, Moskewicz MW, et al. SqueezeNet: alexNet-level accuracy with 50x fewer parameters and <0.5 MB model size. In International Conference on Learning Representations (ICLR). 2017.
- Howard AG, Zhu M, Chen B, et al. Mobilenets: efficient convolutional neural networks for mobile vision applications. arXiv:1704.04861. 2017.
- Zhang X, Zhou X, Lin M, et al. ShuffleNet: an extremely efficient convolutional neural network for mobile devices In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition (CVPR). IEEE; 2018. pp. 6848–6856.
- Sandler M, Howard A, Zhu M, et al. MobileNetV2: Inverted residuals and linear bottlenecks In Proceedings of the IEEE conference on Computer Vision and Pattern Recognition (CVPR). IEEE; 2018. pp. 4510–4520.
- Ma N, Zhang X, Zheng HT, et al. ShuffleNet V2: practical guidelines for efficient CNN architecture design In Proceedings of the European Conference on Computer Vision (ECCV). 2018. pp. 116–131.
- Howard A, Sandler M, Chu G, et al. Searching for MobileNetV3. In Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV). 2019. pp. 1314–1324.
- Tan M, Le QV. EfficientNet: rethinking model scaling for convolutional neural networks. In International Conference on Machine Learning. PMLR 97. 2019. pp. 6105–6114.
- Valanarasu JMJ, Oza P, Hacihaliloglu I, et al. Medical transformer: gated axial-attention for medical image segmentation. In Proceedings of Medical Image Computing and Computer Assisted Intervention, LNCS. vol. 12091. 2021. p. 36–46.
- Hong W-Y, Kao C-L, Kuo Y-H, et al. CholecSeg8k: a semantic segmentation dataset for laparoscopic cholecystectomy based on Cholec80. arXiv:2021.12453. 2020.
- Valanarasu JMJ, Patel VM. UNeXt: MLP-based rapid medical image segmentation network. In Proceedings of Medical Image Computing and Computer Assisted Intervention, LNCS, vol.13435. 2022. p. 23–33.
- Twinanda AP, Shehata S, Mutter D, et al. EndoNet: a deep architecture for recognition tasks on laparoscopic videos. IEEE Trans Med Imaging. 2017;36(1):86–97. doi: 10.1109/TMI.2016.2593957.
- Carstens M, Rinner FM, Bodenstedt S, et al. The dresden surgical anatomy dataset for abdominal organ segmentation in surgical data science. Sci Data. 2023;10(1):3. doi: 10.1038/s41597-022-01719-2.
- Roß T, Reinke A, Full PM, et al. Comparative validation of multi-instance instrument segmentation in endoscopy: results of the ROBUST-MIS 2019 challenge. Med Image Anal. 2021;70:101920. doi: 10.1016/j.media.2020.101920.
- Kirillov A, Mintun E, Ravi N, et al. Segment anything. In ICCV. 2023. pp. 4015–4026.
- Ma J, He Y, Li F, et al. Segment anything in medical images. Nat Commun. 2024;15(1):654. doi: 10.1038/s41467-024-44824-z.
- Bian C, Yuan C, Ma K, et al. Domain adaptation meets zero-shot learning: an annotation-efficient approach to multi-modality medical image segmentation. IEEE Trans Med Imaging. 2021;41(5):1043–1056. doi: 10.1109/TMI.2021.3131245.