Abstract
Context
A significant number of guidance documents and reporting checklists have been published to support researchers in planning, doing, and writing up scientifically rigorous systematic reviews (SRs). However, compliance of researchers with SR guidance and reporting checklists remains a significant challenge, with the majority of published SRs lacking in one or more aspects of the rigour of methods and transparency of reporting.
Objective
To explore how bench protocol development platforms might be repurposed for improving compliance of SRs with conduct guidance and reporting checklists.
System design
We developed a proof-of-concept technology stack based around a general-purpose, guidance- and checklist-compliant SR protocol that was built in protocols.io. We used the protocols.io platform to create an integrated research planning and data collection process for planning guidance-compliant SRs. We used our own custom code and the mustache templating language to automatically create checklist-compliant first-draft SR protocol documents in Microsoft Word
Discussion
Creating the operational process for SR protocol planning and the technology stack for automated documentation allowed us to develop our theoretical understanding of how such a system may improve compliance with research conduct and reporting standards. This includes the potential value of algorithmic rather than heuristic approaches to conducting and reporting research studies, positioning of labelled data rather than a study manuscript as the primary product of the research process, and viewing the process of developing research standards as being analogous to the development of open software. Our study also allowed us to identify a number of technological issues that will need to be addressed to enable further development and testing of our proposed approach. These include limitations in templating language, especially when working in Microsoft Word, and the need for more data labelling and export formats from protocols.io.
Introduction
A number of conduct guidelines and reporting checklists have been published to support researchers in planning, doing, and writing up scientifically rigorous systematic reviews (SRs). However, compliance with SR conduct guidelines and reporting checklists appears to be a significant challenge for researchers, with consistent evidence of shortcomings in the methodological rigour and comprehensiveness of documentation of SRs from a range of research fields including healthcare (Gao et al. Citation2020; Page et al. Citation2016), environmental science (Pullin et al. Citation2022), socioeconomics (Wang et al. Citation2021), and human environmental health (Menon, Struijs, and Whaley Citation2022). This is potentially a significant source of research waste or even societal harm, with significant resources being used to conduct SRs that may be redundant, present inaccurate findings, or both (Ioannidis Citation2016).
Conduct guidelines specify what an SR ought to do, in order to attain a certain level of scientific quality, with ‘quality’ generally understood as some desired combination of usefulness, credibility (e.g., by reducing bias), and transparency (Whaley, Aiassa, et al. Citation2020). SR guidelines tend to be lengthy and complex. The Institute of Medicine guidance on SRs consists of 82 performance elements and extensive narrative text (Institute of Medicine Citation2011). The Cochrane MECIR standard consists of over 75 performance elements accompanied by a handbook of over 700 pages (Higgins Citation2019; Higgins et al. Citation2019). The COSTER recommendations for SRs of environmental health research of consists of 70 elements (Whaley, Aiassa, et al. Citation2020). The length and complexity of guidance is probably unavoidable when formalising the necessary components of a research process that is in itself lengthy and complex. Nonetheless, such complexity presents a considerable organisational challenge for authors seeking to comply with an SR guideline. This challenge requires a significant level of experience to overcome, to understand the specialist vocabulary used in guidance documents and the often complex concepts that terms in the vocabulary may signify, and the unmentioned actions and practices (relating to e.g., resourcing, management, training, and documentation) that need to be undertaken to comply with a superficially simple recommendation or requirement. Nuance and cultural knowledge of this type is challenging, if not impossible, for a guidance document of finite length to provide, or at least for a user of guidance to successfully internalise.
Reporting checklists are intended to support authors in providing enough information about what they did and found in the course of conducting an SR that a reader can follow their methods and appraise the credibility of their results (Moher et al. Citation2014). As for conduct guidelines, we believe there is a high experience requirement for their successful use. One aspect of this experience requirement is the necessity for users and writers of a checklist to share the same understanding of how terms describing checklist items map onto the processes of a systematic review. Ways in which we have observed this breaking down in a PRISMA report (Page et al. Citation2021), for example, include the misidentification of risk of bias with other study appraisal methods, and the conflation of the Population-Exposure-Comparator-Outcome (PECO) statement for describing the objectives of a SR with PECO-derived eligibility criteria for a SR (Menon, Struijs, and Whaley Citation2022). Further complicating things, each item in a reporting checklist implies a certain set of recommended practices that may not be explicit and might not have been followed by the research team using the checklist (for example, reporting a valid search may, in terms of implied guidance, require engaging with an information specialist).
Proficiency in the technical jargon and implied practices of systematic review requires a potentially lengthy period of acculturation in relevant, competent communities of practice. Seen in those terms, successful use of guidance and checklists like MECIR and PRISMA could be as much an indicator of successful acculturation to a systematic review community (or specifically, MECIR- or PRISMA-centric subcommunities) as they are a facilitator of writing up rigorous SR manuscripts. We speculate that the experience requirement for successful compliance with guidance and checklists could be lowered, if the content of guidance and checklists can be reconfigured in the following way: (1) by interpreting on behalf of a user the recommendations of a conduct standard into a sequence of task-list style operational steps that can be followed by a research team; and (2) by enabling the user to record what is done in each operational step in such a way that the data can later be automatically used for a given reporting task.
A recent technological innovation that could allow for this reduction in experience requirement is provided by bench protocol platforms such as protocols.io (Teytelman et al. Citation2016), that facilitate ‘detailing, sharing, and discussing molecular and computational protocols’. Bench protocol platforms have not been directly designed for automating compliance with complex research guidance and reporting checklists. However, they have several features including detailed recording of operational procedures, data capture when following those procedures, and data export functions that allow captured data to be reused, that suggest they could be appropriated for this purpose.
To better understand how bench protocol tools can be repurposed for complex research design and documentation tasks, we set out to build a proof-of-concept general-purpose operational template for planning and reporting a human environmental health SR. The template is designed to have the following features:
Be an explicit, step-by-step sequence of operational procedures for developing and documenting a human environmental health SR protocol (‘operational procedure’);
Map how each step in the template’s operational procedure translates into compliance with a selected set of conduct standards and reporting checklists (‘compliance mapping’);
Have as output structured, labelled data, to render machine-readable the operational procedures and data outputs of the protocol (‘data labelling’);
Automated assembly from the labelled data a standards- and checklist- compliant draft SR protocol document (‘automated documentation’).
We emphasise that this is a proof-of-concept project. As far as we are aware, a general-purpose operational protocol has not been developed for SRs, nor has automated documentation from data gathered during SR protocol development been demonstrated. As a proof of concept, our technology stack is a prototype and the code has not been finessed. Our target audiences in this paper are tool developers seeking to support SR authors in being standards-compliant, and people working in research policy who are interested in what such tools might look like and how they might be expected to work. While we have endeavoured to create something that could be useful in current form, the general-purpose protocol primarily exists for testing and further development of ideas. We cover various aspects of the limitations and development needs of our approach in the Discussion section, below.
System design
Operational procedure
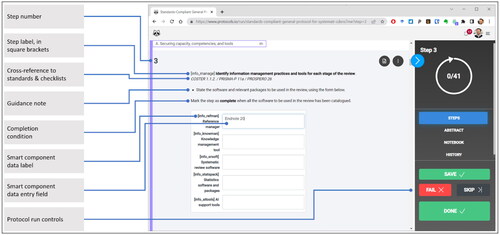
For the general-purpose protocol we revived an experimental build of a step-by-step operationalisation of the COSTER recommendations for conducting environmental health systematic reviews. The experimental build was created in the protocols.io bench protocol platform (Whaley Citation2020; protocols.io accessed 7 July 2022). COSTER is an adaptation for human environmental health SRs of Cochrane’s MECIR standard (Whaley, Aiassa, et al. Citation2020). To build the general-purpose protocol, PW abstracted from COSTER all recommendations relevant to the planning of an SR and interpreted them into a sequence of 41 explicit operational steps. The operational steps include instructions on data that should be collected during each step and the conditions under which each step should be marked as complete ().
Compliance mapping
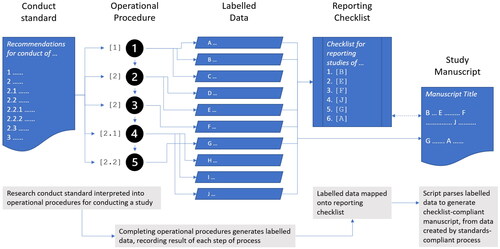
PW cross-referenced each operational step against data reporting requirements for the reporting checklists PRISMA-P (Moher et al. Citation2015), PRISMA-S (Rethlefsen et al. Citation2021), ROSES for SR protocols (Haddaway et al. Citation2018), and PROSPERO registrations (Booth et al. Citation2012), to map how completion of each step contributes to fulfilling the reporting requirements of several commonly-used protocol reporting checklists.
Data labelling
PW created a hierarchical schema for labelling the data collected by completing each step of the general-purpose protocol. The first-level data labels and cross-walk with existing SR guidance and reporting checklists is shown in . The complete labelling schema is available at https://osf.io/vw4e3.
Table 1. Cross-walk of the top-level data labels and steps of the general-purpose protocol with items in the conduct standards and reporting checklists with which the protocol is designed to facilitate compliance.
The labelled data is of two types. For brief, structured data, the user inputs data into the ‘smart component’ feature of protocols.io (shown in ). For complex, unstructured data, such as contextual justification for the SR, the user creates an MS Word document, gives the document a specific name as per protocol instructions, and adds it to an OSF archive. (This is a workaround for limits on data entry in the version of protocols.io we were using for the project.) Instructions for creating an archive, naming the documents, and a link to an OSF template archive with pre-named template documents are included in the general protocol.
Automated documentation
The process of following all the steps of the general-purpose protocol through to completion is called in protocols.io a ‘run’. A record of a run of our general-purpose protocols consists of the following data: the instance of the run in protocols.io itself, that can be exported as a JSON document; the data entered into the smart components, that is included in the protocols.io JSON export (the structured data); and the collection of documents uploaded to the OSF record created by the user during the run (the unstructured data). To automatically convert the run data into a draft protocol document, we created a two step-process ().
Figure 2. The process for automating the documentation of a protocol run. Arrows in blue represent interfaces secured using the Oauth2 authentication standard (https://oauth.net/2/).
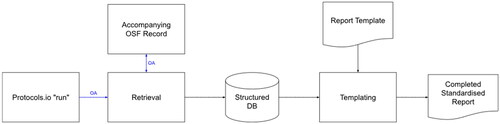
The first stage of the process is retrieval. Here, the run is retrieved from protocols.io using their API (https://www.protocols.io/developers). Arrows in blue in represent interfaces secured using the OAuth2 authentication standard (https://oauth.net/2/). The current build of Protocols.io (accessed 7 July 2022) returns a JSON document that consists of both data entered by the researcher, and other data including formatting instructions for online display. We therefore had to write code to extract individual fields from the protocol run via reference to structural features of the text (e.g., specific punctuation and text block positions). The fields are assigned keys in a key-value store based on the referenced standards to which they apply and the labels assigned to the step using square brackets. For example, the entry ‘Endnote 20’ for reference manager would be stored under the key ‘info_manage’ (label). For the entries that contain references to documents stored on the OSF archive created during the protocol run, we retrieve the OSF record using their API (https://developer.osf.io/). This yields a set of documents in Microsoft Word .docx format that correspond to stages mentioned in the protocol run. We export the index to JSON and store the documents separately on disk for later use. A sample of our JSON export is shown in .
The second stage of the automated documentation process takes these data as input and reassembles them according to a template (an example template can be found at https://osf.io/nckwu). This supports two templating languages. The first is a simple subset of ‘mustache’ markup, a widely-used templating language (https://mustache.github.io/). Here, entries from the JSON document representing structured data from the protocols.io run can be inserted into free text. For example, the entry ‘{{info_refman}}’ is resolved to ‘Endnote 20’, as per the example in . The second, an unnamed proprietary template language, allows template authors to include the contents of an external .docx file completely within the target document, allowing them to concatenate OSF records in any order. The output of this templating process is a Microsoft Word document. In our case, this is a complete summary of all data from the run, intended to be edited by the reviewer into a SR protocol manuscript; however, templates for summarising run data into a PRISMA report or other structured document could also be created.
Anticipated results of running protocol
The general-purpose protocol is available at https://doi.org/10.17504/protocols.io.n92ldydzxl5b/v3. A run of the general-purpose protocol is anticipated to yield three primary outputs:
A comprehensive record of key data that is needed for evaluating the extent to which an SR protocol is compliant with five important guidance and reporting checklists. This data is generated while the user completes each step of the operationalised general protocol, rather than retrospectively after a protocol development process has been completed.
Labelled SR protocol data that, using templates, can in principle be reassembled into any protocol documentation format, e.g., a conference abstract, PROSPERO registration, a protocol manuscript, a PRISMA report, a response to a data sharing request, etc.
After running the TARPD code, a draft SR protocol manuscript. Because the draft manuscript is generated by merging into a fixed template, significant editing will be required before it could be considered publishable. However, since all necessary information is approximately correctly located under relevant subheadings, all required information is present and can be rearranged, with no information omitted.
An example run consisting of a PDF showing what data was input during a test run of the general-purpose protocol, the processed JSON output from the test run, and an automatically generated .docx template created from the test run, are available at https://osf.io/m35px/ (‘Protocol publication’ section). Code for the JSON conversion and .docx document generation is available at https://github.com/StephenWattam/TARPD.
Discussion
Comparison to similar tools
We did not do a systematic review of existing tools for this project, but we did search for similar initiatives from a number of sources. PW screened all GitHub records tagged as ‘systematic-review’, ‘systematic-reviews’, or ‘systematic-literature-reviews’, the first 10 pages of results of a Google search for ‘automated reporting of systematic reviews’, the first 100 results sorted by relevance for searching ‘systematic review’ on the protocols.io platform, the protocol and reporting tools indexed in the Systematic Review Toolbox (http://systematicreviewtools.com/), and the SR support tools reviewed by Kohl et al. (Citation2018). PW also followed up on word-of-mouth recommendations received from discussion of this project with colleagues. For any tool that looked potentially similar to the present project, PW checked public documentation of tool features for information about its functionality. Given the limited documentation that often accompanies tools, we note that our descriptions of a tool may not be a true reflection of the tool’s actual feature set.
The majority of GitHub records seemed to consist of document classifiers designed to support the screening of search results for an SR, along with some tools to support the development of search strategies. We did not find anything obviously related to facilitating compliance with guidance or reporting checklists. While protocols.io has been used as a repository for SR protocols, no SRs appear to have been constructed as operational procedures in the platform. Methods-Wizard from the SR-Accelerator (Clark et al. Citation2020) does have features that generate methods text from input data, but it does not seem to generate labelled data that is designed to be machine-readable, and it does not obviously operationalise the step-by-step process of developing an SR protocol. We did note the existence of a tool for reporting trials in behavioural sciences, the Paper Authoring Tool (West Citation2020), that generates labelled data, has features for automated drafting of a manuscript, and covers some aspects of standards compliance (although without explicit cross-walking). It was not clear if the Paper Authoring Tool is intended to be completed retrospectively, or is a tool that also operationalises the process of planning a study in a way that facilitates compliance with conduct standards.
Cochrane’s RevMan software for conducting SRs of health research (Cochrane Citation2020) appears to be the closest relative of our general-purpose protocol (desktop version 5.4.1, released 21 September 2020). RevMan connects components of a SR protocol or final report to the individual requirements of the MECIR standards, which in turn directs authors to detailed methodological guidance in the relevant sections of the Cochrane Handbook. However, RevMan is a reporting-focused tool that leaves implicit the step-by-step procedures one would follow as a researcher that would result in compliance with MECIR. For example, RevMan does not as an explicit step ask a research team to designate a librarian or information specialist as part of an operational process that increases the effectiveness of search strategies for a review. Also, while some aspects of reporting are automated by the simple provision in RevMan of a structured template designed around PRISMA compliance, authors still manually enter information retrospectively based on their own separate documentation of their process, rather than the SR report being directly generated from data immediately recorded during each operational step. Finally, RevMan is optimised for one SR reporting task (writing a Cochrane review) rather than designed to generate labelled data that can easily be repurposed for any SR reporting task.
Overall, we did not identify a tool that does all three tasks of interpreting an SR protocol into a series of operational steps linked explicitly to SR conduct standards and reporting checklists, collecting structured labelled data in completing those steps, and then automatically generating draft documentation using that data.
Potential strengths of our approach
We believe our approach to a general-purpose SR protocol has three interrelated potential advantages that can be further explored and developed. These are: an emphasis on templated, algorithmic approaches to conducting and reporting research rather than heuristic approaches; taking a view of conduct standards and reporting checklists as being analogous to open-source software; and the positioning of labelled data rather than a study manuscript as the primary output of the research process. We discuss these briefly below.
Algorithmic over heuristic approaches to conducting research
Algorithms are a method for solving a problem in a finite number of steps via the application of a set of formally-defined processes or rules. Heuristics are practical methods for problem-solving that are derived from previous experiences with similar problems. Our opinion is that systematic review is a study design that tends to be conducted heuristically, but could be made more accessible to researchers if formalised to be more algorithmic. Our general-purpose protocol is an attempt to do this.
We note that our approach is algorithmic more in the sense of a detailed recipe in a cook-book than in the more formal sense of being computable. A recipe formalises a sequence of inputs (ingredients) and steps that need to be followed in order to generate an output (a chocolate drip cake). The purpose of the recipe is to transfer expertise between cooks without them having to physically meet, one to directly train another, or a cook having to intuit the recipe from the ingredients and whatever cake-making experience they may have previously acquired. Successfully following the recipe still requires experience and experimentation, but the purpose of the recipe is to reduce that requirement: it means cooks don’t have to remember or internalise a full set of complex rules, making the possibility of successfully creating the final dish more accessible.
Algorithmic protocols could similarly help with standard and checklist compliance. In codifying a set of recommended practices and the recording of data outputs from following them, compliance can become a by-product of following the algorithm rather than something additional that a researcher has to factor into the conduct and documentation of a study. This may help address the problem of consistent under-reporting of key methodological information in SR protocol registrations (Booth et al. Citation2020) and, if extended beyond SR, also for primary studies (Macleod et al. Citation2021).
Guidance and checklists as open-source software
Our approach makes it more transparent how the recommendations and requirements of one or more conduct guidelines, or the requirements that are implicit in one or more reporting checklists, have been interpreted into an operational procedure. Unlike static documents such as the PRISMA checklist (Page et al. Citation2021), we built our procedure in a platform that emulates some aspects of software development. Protocols.io implements several GitHub-like collaboration features (https://github.com/) that allow users of the general protocol to independently build on and develop our prototype. These include allowing users to post questions and comments on each step of the protocol (), and to fork protocols for independent development. Forking is particularly important for tracking versions of protocols.
Figure 4. An example of how a user can comment on a step of the protocol to suggest a feature or improvement. A user could also generate a fork of the protocol to add steps specific to their own requirements.

Once an operational procedure has been written up and shared, it can be discussed, tested, and finessed by those who are using it. Returning to our recipe analogy, cooks will modify recipes for working in environments where an ingredient may need to be substituted for dietary requirements, lack of availability, or for different tastes. Processes may be adapted for different circumstances or levels of skill that may make some culinary techniques unrealistic to implement. This is authoring, adapting, and versioning of the algorithm according to need.
Presenting the variations on a platform where relationships between versions are tracked helps make adaptations discoverable and evaluable, and allows the evolution of processes to be observed. It also implies an approach to standardisation that is more bottom-up and directly responsive to the needs of the user community than the approaches traditionally recommended for developing standards and checklists, such as the review-workshop-document process of Moher et al. (Citation2014). We anticipate this resulting in clouds of related micro-standards adapted around the specific needs of individual research communities. While this might sound chaotic to some, forking and versioning may allow adaptations of standards to be more transparent than the often unclear ways in which users currently interpret compliance with e.g., the PRISMA checklist.
Labelled data as primary, manuscripts as derivative
Our approach positions labelled data as the primary output of the process of conducting a research activity, with documentation as a secondary output that is derived from labelled data. This reconfigures the conventional flow of data reuse in research, whereby researchers generate data, describe the methods for acquiring and how they interpret their data in largely unstructured form via a published document, from which other researchers have to abstract the data they need for reproducing or extending the original study.
When data, including data about methods, is labelled, compliance with a reporting checklist can become a matter of running code that generates a compliant manuscript, rather than retrospectively (and usually manually) cross-checking a document against a complex but routine set of reporting requirements. This kind of cross-checking is the sort of detailed but repetitive task that should be automated. Labelled data is also interoperable: code can be written to generate reports that are consistently compliant with one or more different reporting checklists. Availability of such code should reduce the requirement for an author of an SR protocol to be experienced in any particular reporting format in order for them to produce a comprehensive, checklist-compliant report (illustrated in ).
Current limitations and future development
Technical issues
The proof-of-concept system described above is limited by a number of technical factors in our technology stack that need to be addressed via changes to the stack and/or development of the components within it.
Firstly is the difficulty in processing Microsoft Word documents in Python. Library support for templating, reading, and writing .docx files exists and is mature, but the feature set is limited. We found that existing libraries were not able to apply templates reliably, nor to inspect documents to separately extract text and tables in-order. This limited our templating features to the basic inclusion of fields within running text, or concatenation of full documents (for example, documents can only be concatenated by appending them to the end of a manuscript rather than inserting them in the correct place; while they are in the correct order, the user needs to cut and paste to the correct location). It also makes it easy to break our code, as it looks for text features which if in a different place will not be found. Updating our protocol therefore has to be done with care to preserve structure, which carries implicit rules and is therefore more likely to break.
Secondly, it is apparent that the format used by protocols.io to deliver data over their API is primarily aimed at displaying such data via a web browser, that is, the content is difficult to separate from its formatting. OSF records exhibit a similar issue: though every object within the OSF system has a unique ID that makes referencing simple in principle, this ID is not exposed to the end user except in certain circumstances, meaning it is difficult for users to link directly to documents when completing a protocol run. This could be solved a number of ways, though within our system we relied on unique filenames to identify entries.
Thirdly, data collection features in protocols.io are not yet well-developed. The platform has been designed for recording bench protocols, with data collected mainly for the purpose of process auditing and validation. We appropriated these data collection features for recording data from completion of a step of the general purpose protocol, but with limited success. The number of .docx documents that need to be uploaded to an OSF record is an example of a workaround to address this issue. Allowing users to easily create richer documentation in-situ within the protocol run would be preferable to creating and linking to a series of documents external to the general protocol template.
Mitigating these limitations requires either significant software engineering effort using current technology choices, or changing to a different technology stack. Regarding the latter, we suggest changing the templating format from Word to to HTML, which has a much more mature ecosystem of development tools, and is relatively straightforward to convert to .docx formats if and when required. Other languages and platforms (such as .net) may provide better support for editing such formats.
General development
We only did enough development of our general-purpose protocol to prove the concept. Further operational steps could be added to realise a more complete mapping of conduct and reporting requirements, particularly for PROSPERO where administrative data is lacking from our general protocol, and for item 16 of ROSES that asks for information about validation of search strategy. Nor has our operationalisation been user-tested or optimised. We also think that modularisation of our protocol would be helpful for improving the efficiency of development and testing, and providing opportunities for finessing specific components of the operationalised process independently on an as-needed basis, as per the open software analogy discussed above. For example, we have previously created a protocol for pilot-testing the screening process of a SR (Whaley, Garside, and Eales Citation2020), that could be a module for step 21 of our general-purpose protocol.
While our data labelling allows computational assembly of protocol documentation, our tooling does not yet extend to doing anything computational with the data that has been labelled. Introducing and exploiting semantic authoring features for increased machine-readability and more granular labelling that is integrated with ontologies would be beneficial for a number of reasons (Whaley, Edwards, et al. Citation2020), including interoperability of data between systems and the validation of entered data.
We also note that conduct standards and reporting checklists can be viewed as interventions for improving research quality via the dissemination of written instructions. We believe we have good theoretical reasons for thinking our approach could be a more effective intervention than conventional standards and checklists, but there is not yet any empirical evidence that this is the case. Therefore, there should be research into how effective our approach is, for whom it works best, how much the granularity of operationalisation affects effectiveness, among other ideas. In comparison to conventional approaches, our bottom-up approach to standardisation will have its own challenges in relation to development, evaluation, and validation of the standards that are produced, and the creation of suitable workflows for addressing them in what will be a nebulous and distributed standards ecosystem.
Finally, even if the system is shown to be effective among users, there is no guarantee there will be enough users of the system to make a difference to the general publishing standards such systems seek to improve. We are encouraged by a recent pilot study that suggests researchers may be open to the increased formalisation of the reporting of study data (Wilkins et al. Citation2022), and our approach is built on the assumption that automated research documentation will incentivise the use of operationalised protocols. However, we do not have a well-developed theory based on empirical evidence as to what conditions need to be fulfilled in order for significant uptake of a system such as the one we have prototyped. We subscribe to the view that infrastructure should be viewed broadly as an installed base of both technology and social arrangements, including group membership and conventions of practice (Edwards et al. Citation2009), and that successfully securing uptake of systems analogous to ours will require both to be addressed.
Conclusion
We have presented a general-purpose SR protocol that is intended to serve as a proof-of-concept for improving compliance with SR conduct standards and reporting checklists. We theorised that general-purpose protocols would work by reducing the experience requirement for successfully using SR standards and checklists in a research process. We articulated some of the potential advantages of our approach, including viewing the problem of compliance as benefiting from an algorithm-like solution, treating the development of research standards and checklists as being analogous to open-source software development, and the positioning of labelled data as the primary output of research from which a manuscript is a derivative product. We acknowledge a range of issues with our existing technology stack, and have outlined a plan for further development that includes technical development and the generation of empirical evidence for the effectiveness of our proposed approach.
Open Scholarship
![]()
![]()
This article has earned the Center for Open Science badges for Open Data and Open Materials. The data and materials are openly accessible at https://osf.io/zjevp/ and https://github.com/StephenWattam/TARPD and https://osf.io/zjevp/. To obtain the author’s disclosure form, please contact the Editor.
Authors contributions
Created using Tenzing app (Holcombe et al. Citation2020).
Conceptualization: Paul Whaley. Data curation: Stephen Wattam. Funding acquisition: Paul Whaley and John Vidler. Investigation: Paul Whaley and Stephen Wattam. Methodology: Paul Whaley and Stephen Wattam. Project administration: John Vidler. Software: Stephen Wattam. Supervision: John Vidler. Validation: Stephen Wattam. Visualisation: Paul Whaley and Stephen Wattam. Writing – original draft: Paul Whaley. Writing – review & editing: Paul Whaley, Stephen Wattam, Anna Mae Scott and John Vidler.
Ethics statement
Ethical approval and informed consent were not required for this research, as it involved neither human nor animal participants.
Preregistration
This study was not preregistered.
Acknowledgements
The authors would like to thank: Lancaster University for funding; EBTC’s Scientific Advisory Council for pre-submission review and comments; and all 26 members of the Meta-Data Enhanced Study Templates discussion group, as chaired by PW, for ideas and discussions that inspired or influenced this manuscript, in particular Mónica González-Márquez, Emma Ganley (Protocols.io), Peter Murray-Rust (Cambridge), and Joel Chan (University of Maryland).
Disclosure statement
PW declares the following financial interests: consultancy services for the Evidence-Based Toxicology Collaboration (EBTC) at Johns Hopkins University Bloomberg School of Public Health; Lancaster University, mainly around securing impact of his PhD research (completed 2020); the University of Central Lancashire (UCLAN), which has involved developing furniture fire safety models relevant to reducing the impact of the use of chemical flame retardants in ensuring fire safety for the UK Government Agency OPSS; and for Yordas Group, to deliver training in SR methods for EU JRC. PW also receives an Honorarium as Systematic Reviews Editor for the journal Environment International. In general, PW’s consultancy work is around development and promotion of systematic review methods in environmental health research, delivering training around these methods, and providing editorial services. He is applying for funding to develop evidence surveillance and automated SR documentation methods through Lancaster University, which if successful will arrive within the next two years.
PW also declares the following non-financial interests: a historical, public commitment to high quality systematic review methods in support of environmental health policy and decision-making; active involvement in a range of projects intended to improve research standards in evidence synthesis. These include development of SR conduct standards (e.g., COSTER), reporting checklists (e.g., ROSES), and critical appraisal tools (e.g., CREST_Triage, IV-CAT) to support the production of high quality systematic reviews, evidence maps, and other study designs. PW is an active member of the GRADE Working Group, has a senior role at EBTC that involves promoting SR and other evidence-based methods in toxicology and environmental health, and is a member of an informal UK NGO network promoting good environmental policy in the UK, and several Brussels-based environmental policy advocacy groups including HEAL and EEB. In general, PW has a strong interest in supporting precautionary, evidence-based approaches to environmental health policy.
SW declares providing scientific & technical consultancy services as a subcontractor to PW on a UK OPSS-funded study into the use of chemical fire retardants in furniture. SW has also provided scientific consultancy services in unrelated fields, and states that publication of work in any area impacts chances of future employment.
AMS declares that, as part of her role at Bond University, she provides training in: evidence-based medicine and systematic review methodology (including automation tools), to Australia-based academics, clinicians and students. Those workshops charge a registration fee. AMS is part of the Systematic Review Accelerator team at Bond University, which develops and makes publicly available automation tools to accelerate completion of systematic reviews, as well as conducting research in this area (all tools are available free of charge). AMS is an Associate Editor for Systematic Reviews, a BMC/Springer Nature journal (non-remunerated).
JV is a Senior Research Associate at Lancaster University and declares having no interests that may compromise the integrity of the research described in this manuscript.
Data availability statement
All data is available from the accompanying OSF project record at https://doi.org/10.17605/OSF.IO/ZJEVP and GitHub repository at https://github.com/StephenWattam/TARPD
Additional information
Funding
References
- Booth, A., M. Clarke, G. Dooley, D. Ghersi, D. Moher, M. Petticrew, L. Stewart, et al. 2012. “The Nuts and Bolts of PROSPERO: An International Prospective Register of Systematic Reviews.” Systematic Reviews 1 (1): 1. https://doi.org/10.1186/2046-4053-1-2
- Booth, A., A. S. Mitchell, A. Mott, S. James, S. Cockayne, S. Gascoyne, C. McDaid, et al. 2020. “An Assessment of the Extent to Which the Contents of PROSPERO Records Meet the Systematic Review Protocol Reporting Items in PRISMA-P.” F1000Research 9: 773. https://doi.org/10.12688/f1000research.25181.2
- Clark, J., P. Glasziou, C. Del Mar, A. Bannach-Brown, P. Stehlik, and A. M. Scott. 2020. “A Full Systematic Review Was Completed in 2 Weeks Using Automation Tools: A Case Study.” Journal of Clinical Epidemiology 121: 81–13. https://doi.org/10.1016/j.jclinepi.2020.01.008
- Cochrane. 2020. ReviewManager [Windows]. Available at: https://training.cochrane.org/online-learning/core-software/revman.
- Edwards, P., G. Bowker, S. Jackson, and R. Williams. 2009. “Introduction: An Agenda for Infrastructure Studies.” Journal of the Association for Information Systems 10 (5): 364–374. https://doi.org/10.17705/1jais.00200
- Gao, Y., Y. Cai, K. Yang, M. Liu, S. Shi, J. Chen, Y. Sun, et al. 2020. “Methodological and Reporting Quality in non-Cochrane Systematic Review Updates Could Be Improved: A Comparative Study.” Journal of Clinical Epidemiology 119: 36–46. https://doi.org/10.1016/j.jclinepi.2019.11.012
- Haddaway, N. R., B. Macura, P. Whaley, and A. S. Pullin. 2018. “ROSES Reporting Standards for Systematic Evidence Syntheses: Pro Forma, Flow-Diagram and Descriptive Summary of the Plan and Conduct of Environmental Systematic Reviews and Systematic Maps.” Environmental Evidence 7 (1): 7. https://doi.org/10.1186/s13750-018-0121-7
- Higgins, J. P. T., ed. 2019. Cochrane Handbook for Systematic Reviews of Interventions version 6.0 (updated July 2019). Hoboken: Cochrane. www.training.cochrane.org/handbook.
- Higgins, J. P. T, T. Lasserson, J. Chandler, D. Tovey, and R. Churchill. 2019. Methodological Expectations of Cochrane Intervention Reviews (MECIR). Hoboken: Cochrane. https://community.cochrane.org/mecir-manual.
- Holcombe, A. O., M. Kovacs, F. Aust, and B. Aczel. 2020. “Documenting Contributions to Scholarly Articles Using CRediT and Tenzing.” PLOS One 15 (12): e0244611. https://doi.org/10.1371/journal.pone.0244611
- Institute of Medicine. 2011. Finding What Works in Health Care: Standards for Systematic Reviews. Washington, D.C.: National Academies Press. https://doi.org/10.17226/13059
- Ioannidis, J. P. A. 2016. “The Mass Production of Redundant, Misleading, and Conflicted Systematic Reviews and Meta-Analyses.” The Milbank Quarterly 94 (3): 485–514. https://doi.org/10.1111/1468-0009.12210
- Kohl, C., E. J. McIntosh, S. Unger, N. R. Haddaway, S. Kecke, J. Schiemann, R. Wilhelm, et al. 2018. “Online Tools Supporting the Conduct and Reporting of Systematic Reviews and Systematic Maps: A Case Study on CADIMA and Review of Existing Tools.” Environmental Evidence 7 (1): 1–17. https://doi.org/10.1186/s13750-018-0115-5
- Macleod, M., A. M. Collings, C. Graf, V. Kiermer, D. Mellor, S. Swaminathan, D. Sweet, et al. 2021. “The MDAR (Materials Design Analysis Reporting) Framework for Transparent Reporting in the Life Sciences.” Proceedings of the National Academy of Sciences of the United States of America 118 (17): e2103238118. https://doi.org/10.1073/pnas.2103238118
- Menon, J. M. L., F. Struijs, and P. Whaley. 2022. “The Methodological Rigour of Systematic Reviews in Environmental Health.” Critical Reviews in Toxicology 52 (3): 167–187. https://doi.org/10.1080/10408444.2022.2082917
- Moher, D., D. Altman, I. Simera, and E. Wager. 2014. Guidelines for Reporting Health Research: A User’s Manual. Hoboken: John Wiley & Sons. https://play.google.com/store/books/details?id=ojFEBAAAQBAJ
- Moher, D., L. Shamseer, M. Clarke, D. Ghersi, A. Liberati, M. Petticrew, P. Shekelle, et al. 2015. “Preferred Reporting Items for Systematic Review and Meta-Analysis Protocols (PRISMA-P) 2015 Statement.” Systematic Reviews 4 (1): 1. https://doi.org/10.1186/2046-4053-4-1
- Page, M. J., L. Shamseer, D. G. Altman, J. Tetzlaff, M. Sampson, A. C. Tricco, F. Catalá-López, et al. 2016. “Epidemiology and Reporting Characteristics of Systematic Reviews of Biomedical Research: A Cross-Sectional Study.” PLOS Medicine, 13 (5): e1002028. https://doi.org/10.1371/journal.pmed.1002028
- Page, M. J., J. E. McKenzie, P. M. Bossuyt, I. Boutron, T. C. Hoffmann, C. D. Mulrow, L. Shamseer, et al. 2021. “The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews.” BMJ 372: n71. https://doi.org/10.1136/bmj.n71
- Pullin, A. S., S. H. Cheng, J. D. Jackson, J. Eales, I. Envall, S. J. Fada, G. K. Frampton, et al. 2022. “Standards of Conduct and Reporting in Evidence Syntheses That Could Inform Environmental Policy and Management Decisions.” Environmental Evidence 11 (1): 1–11. https://doi.org/10.1186/s13750-022-00269-9
- Rethlefsen, M. L., S. Kirtley, S. Waffenschmidt, A. P. Ayala, D. Moher, M. J. Page, J. B. Koffel, et al. 2021. “PRISMA-S: An Extension to the PRISMA Statement for Reporting Literature Searches in Systematic Reviews.” Journal of the Medical Library Association 109 (2): 174–200. https://doi.org/10.5195/jmla.2021.962
- Teytelman, L., A. Stoliartchouk, L. Kindler, and B. L.Hurwitz. 2016. “Protocols.io: Virtual Communities for Protocol Development and Discussion.” PLOS Biology 14 (8): e1002538. https://doi.org/10.1371/journal.pbio.1002538
- Wang, X., V. Welch, M. Li, L. Yao, J. Littell, H. Li, N. Yang, et al. 2021. “The Methodological and Reporting Characteristics of Campbell Reviews: A Systematic Review.” Campbell Systematic Reviews 17 (1): e1134. https://doi.org/10.1002/cl2.1134
- West, R. 2020. “An Online Paper Authoring Tool (PAT) to Improve Reporting of, and Synthesis of Evidence from, Trials in Behavioral Sciences.” Health Psychology 39 (9): 846–850. https://doi.org/10.1037/hea0000927
- Whaley, P. 2020. “Generic Protocol for Environmental Health Systematic Reviews Based on COSTER Recommendations v1”, Protocols.io. ZappyLab, Inc. https://doi.org/10.17504/protocols.io.biktkcwn
- Whaley, P., S. W. Edwards, A. Kraft, K. Nyhan, A. Shapiro, S. Watford, S. Wattam, et al. 2020. “Knowledge Organization Systems for Systematic Chemical Assessments.” Environmental Health Perspectives 128 (12): 125001. https://doi.org/10.1289/EHP6994
- Whaley, P., E. Aiassa, C. Beausoleil, A. Beronius, G. Bilotta, A. Boobis, R. de Vries, et al. 2020. “Recommendations for the Conduct of Systematic Reviews in Toxicology and Environmental Health Research (COSTER).” Environment International 143: 105926. https://doi.org/10.1016/j.envint.2020.105926
- Whaley, P., R. Garside, and J. F. Eales. 2020. “A general protocol for pilot-testing the screening stage of a systematic review (manual) v1”, protocols.io ZappyLab, Inc. https://doi.org/10.17504/protocols.io.bkc9ksz6
- Wilkins, A. A., P. Whaley, A. S. Persad, I. L. Druwe, J. S. Lee, M. M. Taylor, A. J. Shapiro, et al. 2022. “Assessing Author Willingness to Enter Study Information into Structured Data Templates as Part of the Manuscript Submission Process: A Pilot Study.” Heliyon 8 (3): e09095. https://doi.org/10.1016/j.heliyon.2022.e09095