MicroRNAs (miRNAs) are a class of highly evolutionarily conserved non-coding RNAs (ncRNAs) that modulate gene expression. They play fundamental roles in the regulation of gene expression by pairing via partial Watson and Crick interactions with complementary sequences within the 3′ untranslated regions (3′UTRs) of targeted transcripts Citation[1] and several studies have shown that the expression of miRNAs is deregulated in human malignancies. For ncRNAs and miRNAs, such gene-profiling studies in tumorigenic tissues have identified significant signatures that are of both diagnostic and prognostic value. The challenge is to translate the numerous known examples linking dysregulated expression of miRNAs to cancer into the development of novel therapeutics.
In this issue, the authors Cho et al. Citation[2] summarize the implications of miRNAs in malignancies and discuss the functions of both oncogenic and tumor suppressive miRNAs. The Expert Opinion represents an important summary of the current field, and the efforts and steps taken to use miRNAs as potential applications in therapy for the treatment of cancers. As discussed earlier Citation[3], the importance of miRNAs in fine-tuning gene expression is highlighted by the demonstration that changes in the abundance of a single miRNA can affect the levels of expression of hundreds of different proteins. Thus, it is not surprising that a single dysregulated miRNA can push cells into a transformed state.
Even nowadays the number of identified miRNAs is high, the number of experimentally validated targets is meager. A single miRNA can target several mRNAs through imperfect base complementarity Citation[4] and bioinformatics data have shown that many miRNAs are highly conserved, which suggests a strong evolutionary pressure. Indeed, miRNA deficiencies or excesses have been linked with a variety of clinically important diseases, among them are viral infections, myocardial infarction, metabolic diseases, neurodegenerative diseases or, as presented in the published article, cancer Citation[5].
Historically, the involvement of miRNAs in cancer was first found in a study on chronic lymphocyte leukemia. A region containing miR-15 and miR-16 at the chromosomal location 13q14 was very often found to be deleted in the majority of chronic leukemia cases Citation[6]. Until now, correlations like this have been found quite frequently and many miRNA are associated with primary human tumors Citation[7]. More than 50% of the human miRNA genes are located at genomic sites implicated in cancers, such as fragile and common breakpoint sites Citation[7] and copy number variations (CNVs) are also frequently found in genomic regions containing miRNAs and miRNA cluster as well as altered expression levels of miRNA genes in transformed cells Citation[8].
The authors Cho et al. Citation[2] have presented several studies demonstrating that miRNAs are involved in numerous pathways in both normal and aberrant cells. For instance, the reduced expression of several miRNA and miRNA cluster is linked with the perturbation of the normal cell-cycle control. For instance, the forced expression of miRNAs, like miR-26a, results in increased numbers of cells in the G1 stage of the cell cycle and fewer cells in the proliferative S phase. Such studies reveal a consistent reduction in the levels of this miRNA in cancer cells versus normal cells; to analyze the therapeutic potential of miR-26a for carcinoma one has to overcome several biological challenges, such as the delivery of miRNA constructs into cancer cells in vivo.
Novel miRNA-based therapeutic approaches target the ncRNAome, including, for instance, miRNA expression levels and improved designs of miRNA mimics or more precise target predictions, prevent off-target effects of novel drugs and make miRNAs potentially a highly efficient class of therapeutics. For miRNA-based therapeutic studies two direct strategies are currently under investigation, viz i) the inhibition of target miRNAs with antisense constructs like antagomiRs or ii) the overexpression of given miRNAs to inhibit the expression of protein-coding genes. Indirect strategies include the use of novel drugs that modulate miRNA expression levels by targeting their processing or transcription directly (). Also, miRNA-based biomarkers have a significant impact on the development of both therapeutic and diagnostic agents, a concept known as theranostics and are highly relevant for drug development and personalized medicine Citation[9].
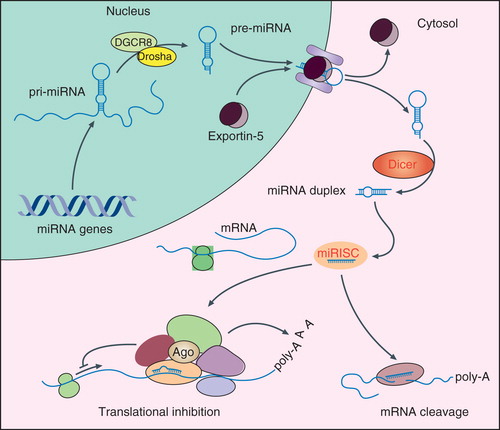
Figure 1. MicroRNA (miRNA) biogenesis. The transcription of miRNA genes by RNA polymerase II (Pol II) results in primary miRNA transcripts (pri-miRNAs), which are then cleaved in the nucleus by a complex of Drosha and DGCR8. The resulting precursor miRNA hairpin (pre-miRNA) is exported into the cytosol in an exportin-5-dependent pathway and further processed by the RNase dicer to an intermediate miRNA duplex. The leading strand is then loaded into the (mi)RNA-induced silencing complex (mi)RISC, whereas the second strand subjected to degradation. The strand selection depends on the thermodynamic characteristics of the miRNA duplex. The (mi)RISC complex is guided to target mRNAs sequences that are located within the 3′ untranslated regions (3′UTRs) of the mRNA. Following these reactions, the mRNA is targeted by translational repression and/or degradation.
The authors give examples of studies that have demonstrated the feasibility of restoring tumor suppressive miRNAs and targeting oncogenic miRNAs in cancer therapy. The greatest hurdles still lie in the systemic delivery of ncRNA-based therapeutics and the stability of the constructs. Also, it has become evident that epigenetic effects play a crucial role in the complexity of human tumorigenesis, as well as DNA methylation patterns and histone code-based gene regulation Citation[10].
In view of novel miRNA-based therapeutics, the activation of miRNAs with so-called agomiRs or the silencing with antagomiRs could probably become such a therapeutic tool and novel strategy for cancer treatment. Both the RNAi technology in general, and the targeted control of gene transcription in particular, have a significant therapeutic potential for novel biomarker-based assays in diagnostics and for the development of novel class of drugs. For ncRNAs, gene-profiling studies in tumorigenic tissues have identified significant signatures that are of both diagnostic and prognostic value. Notwithstanding the encouraging results of studies on miRNAs, many challenges remain. For instance, the expression of miRNAs in tissues is temporal, spatial specific and is influenced by various factors, including epigenetic signals, pathology, infections, etc. As such, it becomes difficult to identify a consistent miRNA signature for diagnostic and prognostic purposes.
Generally speaking, small ncRNAs, and miRNAs in particular, have greatly changed our view of tumorigenesis and comprise a great potential to improve the treatment of malignancies with prospective novel approaches. At present, such novel therapeutics are based on the miRNA–mRNA 3′UTR interaction. However, this view may be too simplistic since several complex miRNA mechanisms and regulatory layers on both the protein and the genomic level control gene regulation, leading back to the question ‘What's regulated in gene regulation?’.
With improved designs of miRNA mimics, more precise target predictions, the prevention of off-target effects, pharmacodynamic and pharmacokinetic studies, distinctive miRNA theranostics will be obtained. As a result, the reprogramming of the ncRNAome or miRNAome of cancer patients might become possible. Also, combined approaches of chemotherapy and novel miRNA-based agents are a new path toward clinical applications. An encouraging example is a study in which miR-128 has been shown to modulate steroid refractoriness in acute lymphocytic leukemia. Thus, a strategy in the treatment of lymphocytic leukemia is the use of synthetic miRNAs in combination with chemotherapy Citation[11]. With respect to the use of oligonucleotide-based miRNA therapeutics, challenges like poor cellular uptake or a low bioavailability need to be further addressed, as well as off-target effects or long-term safety in humans. A means to target such problems is the combination of miRNA-based therapeutic technologies with other forefront technologies, including nanoparticles or polymers. Definitely, miRNA-based approaches bear a great potential for novel therapeutics in cancer therapy and personalized medicine and will find their way into clinical practice.
Declaration of interest
The author states no conflict of interest and has received no payment in preparation of this manuscript.
Bibliography
- Bartel DP. Cell 2009;136:215-33
- Cho MicroRNAs as therapeutic targets and their potential applications in cancer therapy. Expert Opin Ther Targets 2012;16:8
- Baek D, Bartel DP. Nature 2008;455:64-71
- Brennecke J, Stark A, Russell RB, Cohen SM. PLoS Biol 2005;3(3):e85
- Care A, Catalucci D, Felicetti F, Nat Med 2007;13(5):613-18
- Calin GA, Dumitru CD, Shimizu M, Proc Natl Acad Sci USA 2002;99(24):15524-9
- Calin GA, Liu CG, Sevignani C, Proc Natl Acad Sci USA 2004;101(32):11755-60
- Zhang L, Huang J, Yang N, Proc Natl Acad Sci USA 2006;103(24):9136-41
- Iorio MV, Ferracin M, Liu CG, MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005;65:7065-70
- Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin 2010;60:376-92
- Kotani A, Ha D, Hsieh J, MiR-128b is a potent glucocorticoid sensitizer in MLLAF4 acute lymphocytic leukemia cells and exerts cooperative effects with miR-221. Blood 2009;114:4169-78
