Abstract
The Enigma® ML FluAB-RSV assay (Enigma Diagnostics, Porton Down, Salisbury, UK) is a CE-IVD marked multiplex molecular panel for the detection of influenza A, B and respiratory syncytial viruses in nasopharyngeal swabs. The assay runs on the fully automated Enigma ML platform without further specimen manipulation and provides a sample-to-answer result within 95 min. The reported sensitivity and specificity for influenza A are 100% (95% CI: 98.2–100) and 98.3% (95% CI: 95.5–99.4), respectively, for influenza B are 100% (95% CI: 98.2–100) and 98.7% (95% CI: 96–99.6), respectively, and for respiratory syncytial virus are 100% (95% CI: 98.2–100) and 99.4% (95% CI: 97.2–99.9), respectively.
Community-associated respiratory tract infections are a common cause of acute illness in both adults and children, leading to high rates of hospitalizations and lost working days. Upper respiratory tract infections are predominantly caused by viruses, such as rhinovirus, adenovirus and respiratory syncytial virus (RSV), whereas lower respiratory tract infections are frequently caused by bacteria, such as Streptococcus pneumoniae, Haemophilus influenzae, as well as a broad range of viruses including RSV and influenza. Globally, lower respiratory tract infections are the fourth leading cause of death. Influenza and RSV accounted for 507,900 and 253,500 deaths, respectively, in 2010 Citation[1].
During the past few years, an increasing number of rapid diagnostic tests have become available to diagnose bacterial and viral causes of respiratory tract infections Citation[2]. However, despite significant advances in diagnostic techniques, the majority of patients with suspected infections are still managed with empirical antimicrobials rather than more appropriate, directed therapy informed by the identification of the infecting agent Citation[3]. This overuse of antibiotics in both the community and the hospital setting is considered a significant driver for selecting resistance, hence the wider adoption of rapid tests at the point of decision making has the potential to reduce inappropriate treatment dramatically.
Influenza and RSV are two common viral causes of respiratory infections of global significance. They usually present as self-limited, uncomplicated upper respiratory tract infections characterized by an acute febrile illnesses, or are asymptomatic. Nonspecific symptoms may include headache, fever, myalgia, conjunctivitis, rhinorrhoea, pharyngitis, dyspnoea, nonproductive cough, wheeze and nasal congestion. In some patients, particularly children aged <2 years and those with underlying immunosuppression, influenza and RSV may spread to the lower airways causing pneumonia and pneumonitis Citation[4,5]. Infection in these high-risk patient groups is associated with more severe infection, often requiring hospitalization and excess mortality.
The clinical diagnosis of influenza and RSV is especially problematic during periods of endemic prevalence because other respiratory viruses (e.g., adenovirus, parainfluenza viruses and picornaviruses) also circulate during the same period. Some bacterial infections (e.g., Chlamydia, Legionella and Mycoplasma) may also present with similar nonspecific symptoms, and clinical features are a very poor predictor of infectious etiology Citation[6,7].
A study of 3744 patients derived from clinical trials of zanamavir showed that a combination of fever and cough had the best sensitivities of 64% and a specificity of 67% for detecting influenza Citation[8]. A further study in 706 children found that fever was the only sign that independently predicted influenza infection Citation[9]. Thus, clinical findings may help to identify patients with influenza like illness but are poor predictors for confirming or excluding the diagnosis of influenza Citation[10]. Furthermore, mixed infections with two or more respiratory pathogens are not uncommon, occurring in 10–40% of cases and which further complicates both clinical and laboratory diagnosis Citation[11,12].
Respiratory syncytial virus
RSV is an enveloped, single-stranded sRNA virus of the family Paramyxoviridae family Citation[13]. There is an increased incidence in the winter months in temperate climates; this seasonality follows closely that of influenza viruses Citation[14,15].
RSV most commonly affects children and the elderly Citation[16] and has an incubation period of 2 to 8 days Citation[16]. In the USA, RSV is the most common cause of hospitalization in infants Citation[17]. Almost all children are affected by the age of 2 years; however, immunity is incomplete and reinfections are common. Primarily RSV has traditionally been considered a pediatric infection, and its burden on adults is probably under-recognized.
Transmission is by direct contact with either respiratory droplets or the hands of healthcare workers where it can survive for several hours, and several nosocomial outbreaks have been described Citation[17–21]. In transplant patients and young children, infection with RSV can cause serious complications with mortality rates as high as 70–100% Citation[22,23]. Because of these poor outcomes, most centers will delay stem cell transplant until the patient clears the infection.
Influenza viruses
Influenza is an enveloped, single-stranded RNA virus of the family Orthomyxoviridae. Influenza A is the most common, and it is further subtyped according to the sequence of two glycoproteins: hemagglutinin (H) and neuraminidase (N). Influenza B is less common, but it still causes significant numbers of infection. Influenza C is rare and not usually included in diagnostic assays Citation[5].
In temperate climates, flu causes seasonal infections in the colder winter months. Infection is highly contagious and spread from person to person by droplet infection through sneezing, coughing and by touching inanimate surfaces Citation[24].
Approximately 20% of all children and 5% of all adults globally have symptomatic influenza infection each year Citation[5]. The incubation period is between 1 and 4 days Citation[4]. Each annual flu season is normally associated with a major influenza subtype, which changes from year to year. Influenza is one of the most changeable and mutable viruses affecting humans because of the low fidelity of the viral RNA replicase enzyme and the segmented RNA genome Citation[25]. Influenza A has the ability to undergo antigenic shift causing unpredictable pandemics.
Antiviral drugs, such as oseltamivir and zanamivir, reduce the duration and frequency of symptoms, hospitalizations and complications, such as otitis media, bronchitis and pneumonia, if administered within 48 h and may also reduce mortality in certain populations Citation[26–31]. In addition, using either drug as prophylaxis reduces the risk of developing symptomatic influenza in vulnerable household and hospital contacts.
Conventional testing for influenza & RSV
Accurate diagnosis is essential for the clinical management of the infected patient and of vulnerable healthcare and household contacts because antiviral prophylaxis may be offered to these individuals. The narrow time period available to offer these treatments requires a rapid diagnostic turnaround time, which is a challenge for current conventional testing techniques. Confirmatory laboratory results may influence a range of clinical management decisions including the institution or withholding of antivirals and antibiotics, whether to admit or discharge the patient, and the institution or withholding of infection control precautions. Empirical treatment and presumptive isolation prior to laboratory diagnosis are wasteful of resources and could be contributing to the emergence of resistance. Thus, there is a clinical need for rapid, accurate diagnostics, which could have a real role in helping to prevent indiscriminate and inappropriate use of these agents.
Conventional methods used to diagnose influenza and RSV include culture, direct fluorescent antibody assays and serology. Each of these methods has its disadvantages; slower culture growth, takes 7–14 days, meaning that the result is of little relevance to clinical treatment decisions and infection control interventions Citation[32,33]. In addition, culture requires specialized laboratory facilities and expertise and is reliant on the subjective determination of cytopathic effect. Although highly specific, the direct fluorescent antibody tests are relatively complex, requiring significant expertise and reagents with a turnaround time of 2–4 h. Similarly, serology can offer only a retrospective diagnosis limiting the impact on patient care because antibody titers peak at 4–7 weeks after infection. This technique is most usefully used for conducting seroepidemiological studies Citation[34].
Rapid influenza tests
The two main bottlenecks for conventional laboratory-based testing are usually sample transportation from the patient to the centralized laboratory and the requirement for testing in batches. Rapid antigen tests were introduced to alleviate these delays and move the test closer to the patient Citation[35].
These rapid tests and simple to use tests are enzyme and optical immunoassays for virus antigen. They generally provide results in 15–30 min, and there are a number of commercially available tests for both influenza and RSV Citation[34,36–39], some of which are Committee for Laboratory Improvement Act waived for use as point-of-care tests.
Although these tests are usually fairly inexpensive (US$ 15–20) Citation[40], they are variable in terms of diagnostic accuracy with reported sensitivities ranging from 10 to 80% Citation[41–44]. In a pooled analysis of 159 studies of rapid influenza tests, sensitivity was reported at 62.3% (95% CI: 57.9–66.6%) with a range between 4.4 and 100% and specificity 98.2 (95% CI: 97.5–98.7) with a range between 50.5 and 100% Citation[45]. The early reports of low sensitivity led to the recommendation that negative test results could not be used to rule out infection confidently and that negative samples should be further tested with more sensitive assays Citation[46,47]. In addition, positive samples also require further testing for subtyping.
Despite this, the high specificities and positive predictive values (PPVs) during peak respiratory virus season allow faster diagnosis and treatment decisions and allow improved patient care by limiting additional and often unnecessary diagnostic tests in these patients. Pediatric studies, in particular, have demonstrated the reduced use of antibiotics and more appropriate use of antivirals after introducing influenza rapid testing Citation[48–52].
Molecular methods
The first description of reverse transcription PCR for the detection of influenza was in 1991 and since then molecular diagnostics have been increasingly developed by Pillet et al. Citation[53–55]. These have replaced traditional viral culture as the preferred diagnostic technique because of problems with turnaround time (usually 3 to 14 days) and reduced sensitivity of other methods Citation[34,56]. In periods of outbreaks, these assays are very useful for testing large amounts of samples in a relatively short time.
Despite significantly quicker turnaround times, most of the molecular assays for detection of influenza and other respiratory viruses must be performed by specialist personnel in a centralized laboratory setting. Most samples will be tested in batches, and there may be significant sample transportation time to include.
Recently, there has been a trend to automation and miniaturization and molecular assays, and there are now several commercially available tests that might be suited to deployment in settings other than centralized laboratories. Committee for Laboratory Improvement Act waived or moderately complex tests can be performed by appropriately trained laboratory assistants or similar healthcare workers on instruments located outside of the centralized laboratory. Examples of this might be rapid response or satellite laboratories, where results can be used to make critical patient management decisions.
Current commercially available assays include the Alere™-influenza A&B assay Citation[57,58], the Enigma ML FluAB-RSV Assay (Enigma Diagnostics, Salisbury, UK), the FilmArray® (BioFire Diagnostics, Inc., Salt Lake City, UT, USA) Citation[59–61], the Liat™ influenza A/B Assay (Roche, Basel, Switzerland), the Simplexa™ Flu A/B & RSV Direct assay (Focus Diagnostics, Cyrpress CA, USA) Citation[62–64], the Verigene® Respiratory Virus Plus test (Nanosphere, Inc., Northbrook, IL, USA) Citation[61,63,65], and the Xpert® Flu assay (Cepheid, Sunnyvale, CA, USA) Citation[66–68]. These assays range from being able to detect one or two targets (Liat and Alere-i) to a multiplex panel of over 20 pathogens including atypical bacteria (FilmArray) and have turnaround times of 15 (Alere-i) to 155 min (Verigene). Relative performance characteristics of these assays are summarized in .
Table 1. Relative performance characteristics of several commercially available, rapid, multiplex molecular assays for the detection of influenza, RSV and other respiratory viruses.
There are multiple laboratory-based diagnostic accuracy studies comparing the performance of several of these assays. These appear to give favorable results, particularly when compared with the rapid enzyme immunoassays Citation[57–68]. However, there are few reports in the literature of examples where these systems have been placed in a near-patient setting. Notable exceptions are a set of two small studies conducted in Japan using the Verigene Respiratory Virus Panel Citation[52,69]. These evaluated the clinical use of this assay in the pediatric outpatient setting and in adult inpatients presenting with influenza like illness, where the assay was performed by non–laboratory-based physicians. The assay was able to facilitate better triage into isolation rooms. A further study evaluated the FilmArray Respiratory Panel when placed in a core laboratory of a regional children’s hospital, this allowed the mean turnaround time to be reduced from 7 to 1.6 h when compared with a direct fluorescence assay performed in a centralized laboratory Citation[70].
Modeling of the use of a rapid influenza PCR in the emergency department has suggested that the economic benefit depends on the prevalence of the disease Citation[71]. At a prevalence of 3 to 7%, it is cost–effective to use a rapid PCR; however, at a prevalence >7%, it is more cost–effective to treat all patients with antivirals. This study assumed a cost of US$ 53 per rapid PCR test, which included labor; clearly, the cost–effectiveness of a less expensive test may dominate. In addition, the model did not consider the often considerable burden of presumptively isolating and using personal protective equipment for patients with a presumptive diagnosis of influenza.
Like many other point-of-care tests that might be used to improve infection prevention and control, there is often a lack of good clinical outcomes data to support their widespread adoption Citation[35]. Although many of these tests might be suited for the use outside of a laboratory setting, their use requires significant training for staff with no molecular biology or even laboratory experience. The use of these tests requires adherence to specific protocols that could potential cause disruption in the clinical workflow and increase demands on already busy clinical personnel. There is a need to study the practicalities of using these tests in a near-patient setting and which look at ease of use, patient and staff acceptability, clinical outcomes and cost–effectiveness.
The Enigma® ML (mini-laboratory) platform
The Enigma ML is a CE-IVD marked, fully integrated and automated molecular testing platform using real-time PCR to produce rapid results in approximately 90 min. It uses single-use disposable cartridges that can be stored at ambient temperature. These are self-contained units containing all reagents necessary for sample extraction, amplification and detection. All assay reagents are retained within the cartridge, minimizing the risk of cross-contamination or human error. The Enigma ML system was designed for ease of use targeting non-laboratory personnel in a point-of-care or near-patient setting.
The system requires less than 2 min of hands on time. Once the patients details have been entered and the cartridge has been loaded, the user can walk away, leaving the instrument to complete testing. A built-in barcode reader facilitates transfer of operator, patient and cartridge/assay identification.
The platform is scalable with each module operating independently; this allows a menu of assays to be run at the same time. This obviates the need for batch testing, allowing true random access and providing clinical results on demand. The instrument has a footprint of 35 × 31 cm for a single module option up to 35 × 94 cm for a six-module option, weighing between 20 and 76 kg. The instrument does not require an external computer because it is controlled by a built-in touch screen color display. shows the platform in a single module configuration.
The Enigma ML FluAB-RSV assay
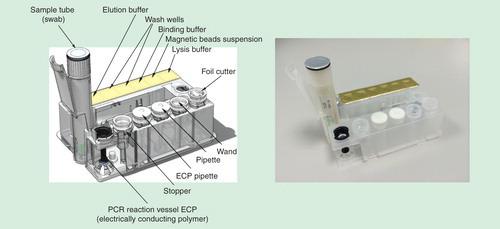
The Enigma ML influenza A/B & RSV A/B assay has CE-IVD designation and is available in all European countries, where CE marking is recognized and there are plans to seek FDA approval in the near future. The assay is based on a magnetic bead sample purification and concentration combined with a fluorogenic reverse transcriptase PCR. Cartridges are foil sealed in blisters for stability and are supplied with a sample collection kit. The nasopharyngeal swab is collected and placed in the sample collection tube, which is then loaded into the cartridge and placed in the ML. As the sample tube is pushed forward, the foil cap is pierced allowing the contents to flow into the cartridge sample well. The sample pipettor transfers and mixes sample into the freeze-dried internal process control (bacteriophage MS2). This validates that both sample preparation and PCR amplification have occurred efficiently. Any failure to detect MS2 indicates that the test has failed. This mixture is then transferred to the lysis well, which contains a guanidine salt. Nucleic acids are released by the combination of chemical and thermal cell lysis. A magnetic wand is used first to transfer magnetic beads, mix the sample and keep the beads suspended. The beads bind the released nucleic acids, which are captured by the wand and transferred to wash wells. Two sequential wash steps are used in this process to remove inhibitory substances before transferring to the elution well where the sample is mixed. After the magnetic beads have been removed, the sample is mixed with the freeze-dried PCR reagents containing fluorescently labeled primers and probes and transferred to the Electrically Conducting Polymer capillary. This is overlayed with mineral oil and a stopper seals the capillary before PCR, which prevents cross-contamination. shows a visual representation of the Enigma ML disposable cartridge.
Primers and probes have been designed for the qualitative detection of influenza A (matrix gene), influenza B (non-structural gene), RSV (fusion gene) together with the previously described internal process control. Primers are used to generate pathogen-specific cDNA for signal detection using one-step reverse transcriptase PCR. Hybridization probes are then used to differentiate between target nucleic acid through dual-hybridization and fluorescent resonant energy transfer. Fluorescent resonant energy transfer allows for a single excitation source, which ‘excites’ the donor fluorophore, this then transfers its energy to an ‘acceptor’ fluorophore when stably positioned in close proximity. The acceptor then emits energy as light at a longer fluorescent wavelength, which is detected in specific channels. The light source and fluorophores that are chosen to ensure only target-specific sequences are detected by the binding of both probes. The amount of acceptor fluorescence is proportional to the amount of PCR product present, which allows for amplification detection.
The ability of ML for high resolution melt enables in silico design of probes, which have specific melting temperatures at which they denature from the amplicon, thereby reducing fluorescence. This allows for differentiation of bound probes, allowing identification of target nucleic acids across the four signaling channels on the ML. These temperature-dependant specific responses are detected and measured by the Enigma ML optics, and an automated algorithmic process reports the presence or absence of each target. Finally, the cartridge is ejected and disposed of as clinical waste.
Performance characteristics in experimental & clinical studies
There are no published data on the clinical performance of the Enigma assay. However, performance data are available from the package insert Citation[72]. The analytical sensitivity claimed by Enigma is 0.2–5 TCID50/ml for influenza A, 0.1–0.5 TCID50/ml for influenza B and 0.06–2.5 TCID50/ml for RSV.
A study was performed using clinical samples obtained in Uganda during the 2013 Southern Hemispehere influenza season. Samples were collected from both adults and children before shipping on dry ice to Enigma Diagnostics for testing. A total of 204 nasopharyngeal swabs derived from patients showing symptoms of influenza-like illness were tested using both the Enigma ML and the Luminex Respiratory Viral Panel FAST version 2 CE-IVD system (Luminex Corp., Austin, TX, USA). Samples were split and tested in parallel, using the xTAG Respiratory Viral Panel as gold standard. For influenza A the sensitivity, specificity, PPV and negative predictive value (NPV) were 100% (95% CI: 98.2–100), 98.3% (95% CI: 95.5–99.4), 89.3% (95% CI: 84.3–92.8) and 100% (95% CI: 98.2–100), respectively. There were three false-positives and no false-negatives. For influenza B, the sensitivity, specificity, PPV and NPV were 100% (95% CI: 98.2–100), 98.7% (95% CI: 96–99.6), 96.4% (95% CI: 92.8–98.2) and 100% (95% CI: 98.2–100), respectively. There were two false-positives and no false-negatives. For RSV, the sensitivity, specificity, PPV and NPV were 100% (95% CI: 98.2–100), 99.4% (95% CI: 97.2–99.9), 96.4% (95% CI: 92.4–100) and 100% (95% CI: 98.2–100), respectively. There was one false-positive and no false-negatives. summarizes the performance characteristics for the three targets.
Table 2. Performance characteristics of the Enigma ML FluAB/RSV assay using the Luminex xTAG RVP as gold standard comparator.
Future developments on the Enigma ML system
Currently, the FluAB-RSV assay is the only available test for the EnigmaML platform; however, Enigma are in late stage development of tests for a multiplexed Respiratory Virus Panel (influenza A and B, RSV, parainfluenza and rhinovirus) and multidrug resistant tuberculosis, which will detect all rifampicin and isoniazid resistant variants directly from sputum, reflecting their initial focus on respiratory infections. In the longer term, Enigma are developing multiplexed tests for sexually transmitted infections (chlamydia gonorrhoea and syphilis), bacterial pneumonia (covering community and ventilator-associated infection), detection of carbapenem-resistant organisms from rectal swabs and a test for bacteremia directly from blood samples.
Conclusion
The Enigma ML FluAB-RSV assay is a new CE-IVD marked assay with encouragingly high sensitivity and specificity. It is rapid, providing a qualitative result in 95 min and allows for operation by nonlaboratory trained personnel. It is thus well suited to deployment in a near-patient setting and is capable of testing up to 30 samples during an 8-h shift. Because of the nonspecific symptoms of respiratory infections, the ability to rule in or out a particular organism rapidly could improve patient pathways and reduce inappropriate prescribing and infection control interventions. The platform and planned menu of assays may be an attractive option for rapid diagnosis in the near-patient setting, as well as centralized laboratories that do not have the expertise or resources to develop their own molecular panels.
Expert commentary
Enigma ML FluAB-RSV assay is a fully automated and integrated test that has the potential to reduce laboratory turnaround times significantly by overcoming delays in sample transportation and batching. The system has the advantage of being able to be run by nonspecifically trained operators close to the frontline.
There are limited clinical data available on this system, and its adoption will depend on results of independently conducted investigations that verify its performance; ideally, these would include more than just diagnostic accuracy data. An assessment of ease of use and clinical use are important factors for healthcare organization to consider before committing to a particular assay.
Five-year view
Fully automated PCR and other molecular tests show great promise in offering sensitive and rapid results that are operator friendly and simple to use. Most are moderate complexity meaning that in North America at least this limits the opportunity for placement in point of care settings, such as emergency departments. However, these are the exact environments where such tests can realize their full potential.
Technological advances will allow increasingly multiplexed assays to be introduced during the next 5 years, permitting whole body system-based syndromic diagnosis of disease, which will enhance epidemiological surveillance. This approach may also be suitable for other areas, such as gastrointestinal infection and meningitis.
Currently available molecular panels are very diverse ranging from the very fast (15 min) detection of one or two targets to a slower but much broader simultaneous diagnosis of a range of both bacterial and viral pathogens. The turnaround time requirement will likely affect the environment in which such a system is placed.
These assays should offer benefits to individual patients in both the hospital and the community setting, as well as enhancing epidemiological surveillance and public health. However, the additional cost and lack of clinical outcomes and cost–effectiveness data will be the major barriers to widespread adoption.
Influenza and respiratory syncytial virus (RSV) are pathogens of global importance with the potential to cause large outbreaks in the community and hospital setting.
Diagnosis based on clinical signs and symptoms alone is unreliable because the features of infection are poorly discriminating. Diagnostic tests are required to confirm the presence of infection.
Rapid influenza antigen tests are poorly sensitive and thus cannot be relied on to exclude infection.
There are a number of commercially available molecular assays that are able to detect influenza and RSV from clinical specimens, many of which are suited to a rapid testing laboratory.
The Enigma® ML FluAB-RSV assay is one such commercially available test, providing a result in 95 min. There are limited data on its performance but early reports suggest high sensitivity and specificity.
There is a need to study the clinical use of these assays, which include data on patient outcomes and cost–effectiveness.
Financial & competing interests disclosure
The authors have received research grants from Enigma Diagnostics Ltd. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the global burden of disease study 2010. Lancet 2012;380(9859):2095-128
- Peaper DR, Landry ML. Rapid diagnosis of influenza: state of the art. Clin Lab Med 2014;34(2):365-85
- Caliendo AM, Gilbert DN, Ginocchio CC, et al. Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis 2013;57(Suppl 3):S139-70
- Cox NJ, Subbarao K. Influenza. Lancet 1999;354(9186):1277-82
- Nicholson KG, Wood JM, Zambon M. Influenza. Lancet 2003;362(9397):1733-45
- Lee N, Qureshi ST. Other viral pneumonias: coronavirus, respiratory syncytial virus, adenovirus, hantavirus. Crit Care Clin 2013;29(4):1045-68
- Huijskens EG, Koopmans M, Palmen FM, et al. The value of signs and symptoms in differentiating between bacterial, viral and mixed aetiology in patients with community-acquired pneumonia. J Med Microbiol 2014;63(Pt 3):441-52
- Monto AS, Gravenstein S, Elliott M, et al. Clinical signs and symptoms predicting influenza infection. Arch Intern Med 2000;160(21):3243-7
- Heinonen S, Peltola V, Silvennoinen H, et al. Signs and symptoms predicting influenza in children: a matched case-control analysis of prospectively collected clinical data. Eur J Clin Microbiol Infect Dis 2012;31(7):1569-74
- Call SA, Vollenweider MA, Hornung CA, et al. Does this patient have influenza? JAMA 2005;293(8):987-97
- Goka EA, Vallely PJ, Mutton KJ, Klaper PE. Single, dual and multiple respiratory virus infections and risk of hospitalization and mortality. Epidemiol Infect 2014;1-11. [Epub ahead of print]
- Karhu J, Ala-Kokko TI, Vuorinen T, et al. Lower respiratory tract virus findings in mechanically ventilated patients with severe community-acquired pneumonia. Clin Infect Dis 2014. [ Epub ahead of print]
- Collins PL, Fearns R, Graham BS. Respiratory syncytial virus: virology, reverse genetics, and pathogenesis of disease. Curr Top Microbiol Immunol 2013;372:3-38
- Stensballe LG, Devasundaram JK, Simoes EA. Respiratory syncytial virus epidemics: the ups and downs of a seasonal virus. Pediatr Infect Dis J 2003;22(2 Suppl):S21-32
- McGuiness CB, Boron ML, Saunders B, et al. Respiratory syncytial virus surveillance in the United States, 2007-2012: results from a national surveillance system. Pediatr Infect Dis J 2014;33(6):589-94
- Sundaram ME, Meece JK, Sifakis F, et al. Medically attended respiratory syncytial virus infections in adults aged ≥ 50 years: clinical characteristics and outcomes. Clin Infect Dis 2014;58(3):342-9
- Zhou H, Thompson WW, Viboud CG, et al. Hospitalizations associated with influenza and respiratory syncytial virus in the United States, 1993-2008. Clin Infect Dis 2012;54(10):1427-36
- Kapikian AZ, Bell JA, Mastrota FM, et al. An outbreak of febrile illness and pneumonia associated with respiratory syncytial virus infection. Am J Hyg 1961;74:234-48
- Chu HY, Englund JA, Podczervinski S, et al. Nosocomial transmission of respiratory syncytial virus in an outpatient cancer center. Biol Blood Marrow Transplant 2014;20(6):844-51
- Meijer A, Overduin P, Hommel D, et al. Outbreak of respiratory syncytial virus infections in a nursing home and possible sources of introduction: the Netherlands, winter 2012/2013. J Am Geriatr Soc 2013;61(12):2230-1
- Singh AK, Jain B, Verma AK, et al. Hospital outbreak of human respiratory syncytial virus (HRSV) illness in immunocompromised hospitalized children during summer. Clin Respir J 2014. [ Epub ahead of print]
- Hertz MI, Englund JA, Snover D, et al. Respiratory syncytial virus-induced acute lung injury in adult patients with bone marrow transplants: a clinical approach and review of the literature. Medicine (Baltimore) 1989;68(5):269-81
- Whimbey E, Champlin RE, Englund JA, et al. Combination therapy with aerosolized ribavirin and intravenous immunoglobulin for respiratory syncytial virus disease in adult bone marrow transplant recipients. Bone Marrow Transplant 1995;16(3):393-9
- Brankston G, Gitterman L, Hirji Z, et al. Transmission of influenza A in human beings. Lancet Infect Dis 2007;7(4):257-65
- Smith EC, Sexton NR, Denison MR. Thinking outside the triangle: replication fidelity of the largest RNA viruses. Annu Rev Virol 2014. [ Epub ahead of print]
- Jefferson T, Jones MA, Doshi P, et al. Neuraminidase inhibitors for preventing and treating influenza in healthy adults and children. Cochrane Database Syst Rev 2014;4:CD008965
- Pebody RG, Harris R, Kafatos G, et al. Use of antiviral drugs to reduce household transmission of pandemic (H1N1) 2009, United Kingdom. Emerg Infect Dis 2011;17(6):990-9
- Muthuri SG, Venkatesan S, Myles PR, et al. Effectiveness of neuraminidase inhibitors in reducing mortality in patients admitted to hospital with influenza A H1N1pdm09 virus infection: a meta-analysis of individual participant data. Lancet Respir Med 2014;2(5):395-404
- Glezen WP. Clinical practice. Prevention and treatment of seasonal influenza. N Engl J Med 2008;359(24):2579-85
- Moscona A. Medical management of influenza infection. Annu Rev Med 2008;59:397-413
- Moscona A. Neuraminidase inhibitors for influenza. N Engl J Med 2005;353(13):1363-73
- Meier CR, Napalkov PN, Wegmüller Y, et al. Population-based study on incidence, risk factors, clinical complications and drug utilisation associated with influenza in the United Kingdom. Eur J Clin Microbiol Infect Dis 2000;19(11):834-42
- Ginocchio CC. Detection of respiratory viruses using non-molecular based methods. J Clin Virol 2007;40(Suppl 1):S11-14
- Kumar S, Henrickson KJ. Update on influenza diagnostics: lessons from the novel H1N1 influenza A pandemic. Clin Microbiol Rev 2012;25(2):344-61
- Moore C. Point-of-care tests for infection control: should rapid testing be in the laboratory or at the front line? J Hosp Infect 2013;85(1):1-7
- Nicholson KG, Abrams KR, Batham S, et al. Randomised controlled trial and health economic evaluation of the impact of diagnostic testing for influenza, respiratory syncytial virus and Streptococcus pneumoniae infection on the management of acute admissions in the elderly and high-risk 18- to 64-year-olds. Health Technol Assess 2014;18(36):1-274
- Peterson S, Dugas AF, Rothman RE. Evaluation of 11 commercially available rapid influenza diagnostic tests–United States, 2011-2012. Ann Emerg Med 2013;61(5):573-7
- Landry ML. Diagnostic tests for influenza infection. Curr Opin Pediatr 2011;23(1):91-7
- Gavin PJ, Thomson RBJr. Review of rapid diagnostic tests for influenza. Clin Appl Immunol Rev 2004;4(3):151-72
- Rapid diagnostic tests for influenza. Med Lett Drugs Ther 1999;41(1068):121-2
- Agoritsas K, Mack K, Bonsu BK, et al. Evaluation of the quidel quickvue test for detection of influenza A and B viruses in the pediatric emergency medicine setting by use of three specimen collection methods. J Clin Microbiol 2006;44(7):2638-41
- Drexler JF, Helmer A, Kirberg H, et al. Poor clinical sensitivity of rapid antigen test for influenza A pandemic (H1N1) 2009 virus. Emerg Infect Dis 2009;15(10):1662-4
- Chen Y, Xu F, Gui X, et al. A rapid test for the detection of influenza A virus including pandemic influenza A/H1N1 2009. J Virol Methods 2010;167(1):100-2
- Bellmann-Weiler R, Beikircher B, Kurz K, et al. Accuracy of bedside antigen tests in the diagnosis of new influenza A/h1n1v infection. Clin Microbiol Infect 2011;17(2):235-7
- Chartrand C, Leeflang MM, Minion J, et al. Accuracy of rapid influenza diagnostic tests: a meta-analysis. Ann Intern Med 2012;156(7):500-11
- Centers for Disease Control and Prevention. Interim recommendations for clinical use of influenza diagnostic tests during the 2009-10 influenza season. 2010. Available from: www.cdc.gov/h1n1flu/guidance/diagnostic_tests.htm
- Centers for Disease Control and Prevention. Rapid diagnostic testing for influenza information for health care professionals. 2010. Available from: www.cdc.gov/flu/professionals/diagnosis/rapidclin.htm
- Ozkaya E, Cambaz N, Coşkun Y, et al. The effect of rapid diagnostic testing for influenza on the reduction of antibiotic use in paediatric emergency department. Acta Paediatr 2009;98(10):1589-92
- Bhavnani D, Phatinawin L, Chantra S, et al. The influence of rapid influenza diagnostic testing on antibiotic prescribing patterns in rural Thailand. Int J Infect Dis 2007;11(4):355-9
- Hojat K, Duppenthaler A, Aebi C. Impact of the availability of an influenza virus rapid antigen test on diagnostic decision making in a pediatric emergency department. Pediatr Emerg Care 2013;29(6):696-8
- Falsey AR, Murata Y, Walsh EE. Impact of rapid diagnosis on management of adults hospitalized with influenza. Arch Intern Med 2007;167(4):354-60
- Nakao A, Hisata K, Matsunaga N, et al. The clinical utility of a near patient care rapid microarray-based diagnostic test for influenza and respiratory syncytial virus infections in the pediatric setting. Diagn Microbiol Infect Dis 2014;78(4):363-7
- Zhang WD, Evans DH. Detection and identification of human influenza viruses by the polymerase chain reaction. J Virol Methods 1991;33(1-2):165-89
- Taubenberger JK, Layne SP. Diagnosis of influenza virus: coming to grips with the molecular era. Mol Diagn 2001;6(4):291-305
- Pillet S, Lardeux M, Dina J, et al. Comparative evaluation of six commercialized multiplex PCR kits for the diagnosis of respiratory infections. PLoS One 2013;8(8):e72174
- Steininger C, Kundi M, Aberle SW, et al. Effectiveness of reverse transcription-PCR, virus isolation, and enzyme-linked immunosorbent assay for diagnosis of influenza A virus infection in different age groups. J Clin Microbiol 2002;40(6):2051-6
- Bell J, Bonner A, Cohen DM, et al. Multicenter clinical evaluation of the novel Alere™ i influenza A&B isothermal nucleic acid amplification test. J Clin Virol 2014;61(1):81-6
- Nie S, Roth R, Stiles J, et al. Evaluation of Alere-i influenza A&B for rapid detection of influenza viruses A and B. J Clin Microbiol 2014;52(9):3339-44
- Babady NE, Mead P, Stiles J, et al. Comparison of the luminex xtag RVP fast assay and the idaho technology filmarray RP assay for detection of respiratory viruses in pediatric patients at a cancer hospital. J Clin Microbiol 2012;50(7):2282-8
- Pierce VM, Elkan M, Leet M, et al. Comparison of the idaho technology filmarray system to real-time PCR for detection of respiratory pathogens in children. J Clin Microbiol 2012;50(2):364-71
- Van Wesenbeeck L, Meeuws H, Van Immerseel A, et al. Comparison of the Filmarray RP, Verigene RV+, and Prodesse Proflu+/FAST+ multiplex platforms for detection of influenza viruses in clinical samples from the 2011-2012 influenza season in Belgium. J Clin Microbiol 2013;51(9):2977-85
- Landry ML, Ferguson D. Comparison of Simplexa™ flu A/B & RSV with cytospin-immunofluorescence and laboratory developed Taqman PCR in predominantly adult hospitalized patients. J Clin Microbiol 2014;52(8):3057-9
- Alby K, Popowitch EB, Miller MB. Comparative evaluation of the Nanosphere Verigene RV+ assay and the Simplexa flu A/B & RSV kit for detection of influenza and respiratory syncytial viruses. J Clin Microbiol 2013;51(1):352-3
- Woodberry MW, Shankar R, Cent A, et al. Comparison of the Simplexa fluA/B & RSV direct assay and laboratory-developed real-time PCR assays for detection of respiratory virus. J Clin Microbiol 2013;51(11):3883-5
- Cho HJ, Jang JW, Ko SY, et al. Evaluation and verification of the Nanosphere Verigene RV+ assay for detection of influenza A/B and H1/H3 subtyping. J Med Virol 2014. [Epub ahead of print]
- Novak-Weekley SM, Marlowe EM, Poulter M, et al. Evaluation of the Cepheid Xpert flu assay for rapid identification and differentiation of influenza A, influenza A 2009 H1N1, and influenza B viruses. J Clin Microbiol 2012;50(5):1704-10
- Salez N, Nougairede A, Ninove L, et al. Xpert flu for point-of-care diagnosis of human influenza in industrialized countries. Expert Rev Mol Diagn 2014;14(4):411-18
- DiMaio MA, Sahoo MK, Waggoner J, Pinsky BA. Comparison of Xpert flu rapid nucleic acid testing with rapid antigen testing for the diagnosis of influenza A and B. J Virol Methods 2012;186(1-2):137-40
- Boku S, Naito T, Murai K, et al. Near point-of-care administration by the attending physician of the rapid influenza antigen detection immunochromatography test and the fully automated respiratory virus nucleic acid test: contribution to patient management. Diagn Microbiol Infect Dis 2013;76(4):445-9
- Xu M, Qin X, Astion ML, et al. Implementation of filmarray respiratory viral panel in a core laboratory improves testing turnaround time and patient care. Am J Clin Pathol 2013;139(1):118-23
- Dugas AF, Coleman S, Gaydos CA, et al. Cost-utility of rapid polymerase chain reaction-based influenza testing for high-risk emergency department patients. Ann Emerg Med 2013;62(1):80-8
- Enigma. Enigma ML FluAB-RSV package insert. Enigma ML