Abstract
The development of the fluorescently labeled tetrameric MHC–peptide complex has enabled the direct visualization, quantification and phenotypic characterization of antigen-specific T cells using flow cytometry and has transformed our understanding of cellular immune responses. The combination of this technology with functional assays provides many new insights into these cells, allowing investigation into their lifecycle, manner of death and effector function. In this article, we hope to provide an overview of the techniques used in the construction of these tetramers, the problems and solutions associated with them, and the methods used in the study of antigen-specific T cells. Understanding how the antigen-specific cells develop and function in different circumstances and with different pathogens will be key to understanding natural host defense, as well as vaccine design and assessment.
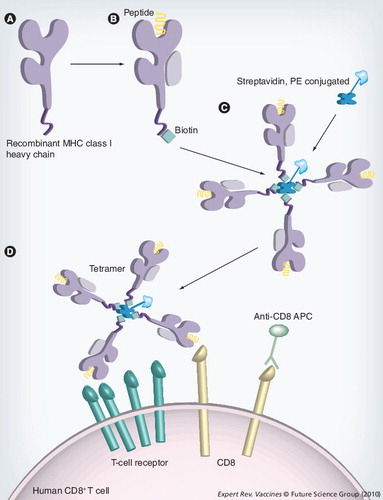
(A) Soluble versions of the heavy-chain MHC class I molecules are synthesized in Escherichia coli. (B) The heavy chain is refolded with β2-microglobulin and the epitope of interest. This refolded MHC molecule is biotinylated with the enzyme BirA at the specific BirA-recognition sequence, at the carboxyl terminus of the MHC molecule. (C)A tetramer is constructed by the addition of four MHC–biotin complexes, binding to a single streptavidin molecule, using the biotin–avidin interaction. This streptavidin molecule is conjugated to a fluorochrome to allow analysis by fluorescence-activated cell sorting. (D) Tetramers are mixed with the cell population that is to be analyzed. The tetramer will bind to CD8+ T cells that are expressing a T-cell receptor that is capable of recognizing the specific peptide and, with the addition monoclonal antibodies, cell populations can then be identified.
APC: Allophycocyanin; PE: Phycoerythrin.
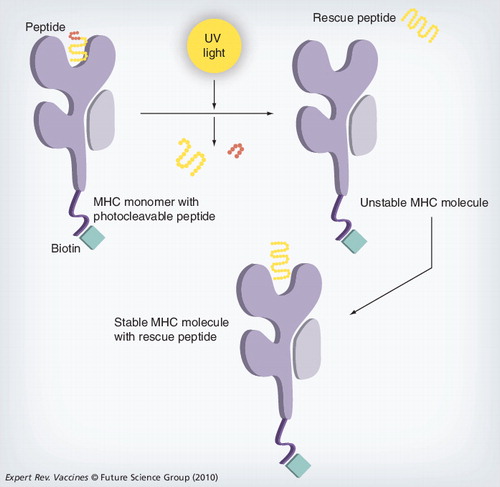
The MHC complex containing the photoliable peptide is irradiated with UV light, cleaving the peptide and producing two fragments that dissociate from the MHC-binding groove. The resulting empty MHC complex is unstable; incubating this MHC with a peptide of interest creates a stable form that can then be used for analysis.
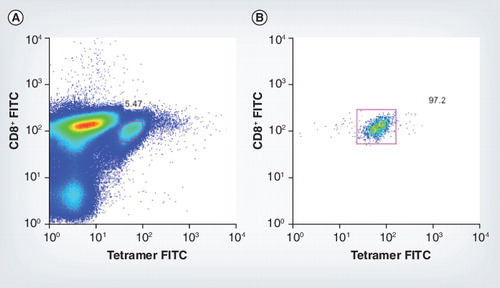
(A) Tetramer-positive mice CD8+ cells for murine cytomegalovirus epitope m45 7 days postinfection, pre-sort. (B) Fluorescence-activated cell sorting plot shows the purity of these tetramer-positive cells post-sort.
FITC: Fluorescein isothiocyanate.
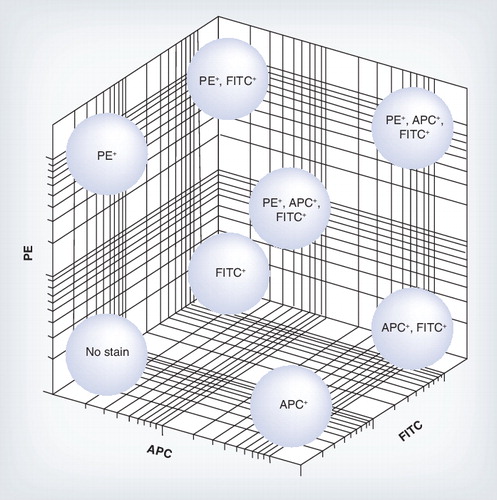
Each tetramer was made by tetramerizing the MHC class I molecule with either one fluorochrome APC, PE and FITC or a mixture, allowing many epitope-specific cell populations to be analyzed from a limited number of fluorochromes.
APC: Allophycocyanin; FITC: Fluorescein isothiocyanate; PE: Phycoerythrin.
Since 1996 there has been a revolution in the characterization of antigen-specific T cells Citation[1]. The catalyst for this revolution was the development of reagents consisting of soluble MHC class I–peptide complexes for the detection of CD8+ cells by flow cytometry. These tetramers provided an efficient way to directly visualize, quantify, phenotype and sort antigen-specific cytotoxic T lymphocytes (CTLs) directly from a wide range of biological samples and have now become standard reagents in laboratories around the world.
The MHC class I molecule is a membrane surface protein found on nearly all cell types. Its function is to bind antigenic peptides and present them on the surface of virally infected or tumor cells. CTLs recognize antigenic peptides, expressed in the MHC class I molecules on the cell surface, via their T-cell receptor (TCR) and, upon recognition, lyse these target cells. Therefore, it is important to measure antigen-specific T cells.
Tetramers have emerged as a key tool for determining the frequency of antigen-specific T cells, particularly in viral infections and after vaccination. They can be combined with other staining techniques to derive detailed information about antigen-specific T cells, such as activation, effector function, proliferation and apoptosis. This has led to an explosion of information on antigen-specific CD8+ T cells and, to a lesser extent, CD4+ T cells.
Staining of T cells relies on the multimerization of the MHC–peptide complexes to overcome the problems raised by the weak monomeric MHC–TCR interactions. The TCR and MHC–peptide complex binding is characterized by the low affinity and fast off rates (disassociation of the MHC complex from the TCR) Citation[2]. This low affinity and fast off rate is required to enable serial contacts of each TCR molecule to bind to multiple MHC–peptide ligands; binding affinities are measured in the micromolar range and off rates are measured in seconds. For this reason, the use of monovalent MHC class I complexes used to stain T cells has generally been unsuccessful. The development of tetramers has allowed multiple MHC–peptide complexes to bind simultaneously to multiple TCRs on the cell surface; this resulted in an increase in the tetramer binding affinity and decreased the off rate, allowing the successful staining of antigen-specific T cells Citation[3]. Indeed tetramers may be subsequently internalized within antigen-specific cells, leading to enhanced staining capacity Citation[4].
Since the introduction of MHC class I tetramers, the technology has been adapted to other complexes recognized by antigen-specific T cells, including MHC class II-restricted CD4+ T cells and glycolipid-specific natural killer (NK) T cells restricted by CD1. These complexes have been used to characterize a range of immune responses in humans, mice, macaques, sooty macaques, chimpanzees, equines, chickens and swine (Box 1)Citation[5–15] and are set to grow as the technology becomes widespread.
Until the recent development of tetramers, the main techniques for analyzing antigen-specific T-cells were CTL assay, limiting dilution assay, intracellular cytokine staining (ICS), enzyme-linked immunospot assay (ELISpot) and surface cytokine capture assays, all of which have their drawbacks. Tetramers allow direct visualization of antigen-specific CTLs without in vitro manipulation by flow cytometry and can be combined with the assays listed allowing functional analysis of T cells, which will be explained in more depth later.
Tetramer construction
MHC class I tetramers
MHC class I–peptide complexes consist of an invariant light chain (β2-microglobulin), a polymorphic heavy chain and an 8–10 amino acid peptide. The peptide forms the essential subunit of the complex as MHC molecules that do not associate with a peptide are unstable and will dissociate. This association is based on specific complementary interactions between the amino acid side chains at the anchor positions of the peptide and the MHC allele-specific pockets.
Classical MHC class I tetramers consist of a complex of four MHC class I glycoproteins loaded with peptide and bound to a fluorescently labeled streptavidin molecule . There are many alternative methods in the construction of MHC class I tetramers but the most common method is by independently expressing the soluble heavy chain of the MHC class I and β2-microglobulin in Escherichia coliCitation[16]. The class I MHC α-chain has the transmembrane region removed, only expressing the extracellular domain and is typically engineered to contain the biotin 15 amino acid recognition tag on the C-terminus. In such preparations, the subunits will be expressed as inclusion bodies within the bacteria.
The heavy chain and the light chain, along with the peptide of interest, are refolded, and the enzyme BirA is used to specifically biotinylate the lysine residue within the 15 amino acid recognition sequence. This reaction can be carried out in vitro or, more recently, has been performed by coexpressing the enzyme along with the MHC protein Citation[17]; in the latter case, the proteins will be biotinylated in vivo and can be used directly. The biotinylated monomer is purified by size-exclusion chromatography (and charge in some settings) and tetramerization is carried out by the addition of streptavidin. A number of different multimerization techniques are now available and the consequent reagents are described as multimers, pentamers, dextramers, and so on Citation[18,19]. In this article, we will use the term tetramer although most of the issues discussed are generic to all forms available.
One of the drawbacks of this technique is that individual tetramers have to be made for each peptide of interest, making it very labor-intensive Citation[16]. A new approach using a peptide exchange method is now available, which allows large collections of tetramers to different antigens to be produced from the one refold Citation[20]. This high-throughput approach involves the production of a MHC class I molecule with a specific peptide that contains a photocleavable moiety; the refold is carried out as normal, producing a stable monomer and, upon irradiation with UV light, the ligand is cleaved and dissociates from the HLA complex . The peptide of interest, which you wish to insert into the MHC molecule, is added to the irradiated mixture replacing the cleaved ligand, thus producing the desired MHC class I molecule with the specific antigen. This method may be used for large-scale T-cell epitope discovery and for T-cell screening that is not feasible by traditional techniques, and can be used in the production of MHC microarrays Citation[21].
MHC class II tetramers
Unlike the relative ease of producing MHC class I tetramers, class II tetramers have been more problematic. Early challenges in the application of tetramer staining CD4+ T cells included not only the difficulties in expression of the allelic variants but also the detection of low-frequency CD4+ T-cell populations and the complications caused by the low T-cell receptor–MHC avidity Citation[5,22]. There have been advances in the technology to overcome these problems, and MHC class II tetramers are in more widespread use and are even commercially available.
MHC class II tetramers can be produced broadly by two methods: the first method is analogous to MHC class I tetramers, by expression of the insoluble denatured subunits in E. coli inclusion bodies, followed by solubilization and in vitro folding in the presence of peptide, purification, biotinylation and tetramerization Citation[5,23]. However, this approach has only been successful at producing a limited number of molecules.
Unlike class I alleles, which are refolded relatively simply in a mild denaturant, class II alleles are complex and may only refold under specific conditions. This complex refolding requires the conditions for each MHC class II protein to be optimized individually for each species and allotype. This complexity may be due to the intrinsic structural instability of soluble class II molecules.
Another method that has been used with more success in producing the class II proteins, and is now probably the most common method, is to coexpress the α- and β-subunits, for example, in mammalian or insect cells Citation[1,24,25]. In some cases, a fusion protein is produced, consisting of peptide fused to the MHC class II β N-terminus via a linker region Citation[22,24]. The covalently attached peptide has access to the peptide-binding site and is able to bind to the peptide-binding groove, stabilizing the construct.
Other modifications available to enhance assembly of the subunits have been developed; these include the use of leucine zippers added to the C-termini of the subunits Citation[26]. Acidic and basic leucine zippers are attached to the extracellular domain of the β- and α-chains of the class II molecules; a linker region is attached, which includes a thrombin cleavage site, and is inserted between the class II chain and leucine zipper and a biotinylation sequence attached to the C-termini of the acidic zipper. This and the use of chimeric IgG Fc domains promote assembly and stability of the heterodimer Citation[25,27,28].
A drawback with the covalently linked peptide is that a new expression vector is required for each MHC–peptide complex; however, by replacing this covalently linked peptide with the class II-associated invariant peptide (CLIP), this problem can be avoided. The CLIP peptide is released through cleavage of the linker region and is replaced with the antigenic peptides of interest in an in vitro peptide exchange reaction Citation[29]. Additionally, HLA-DM, the catalytic peptide exchange factor required for exchange of CLIP in vivo, is used in vitro for antigenic peptide loading of the complex under physiological pH. Empty tetramers (e.g., tetramers not loaded with peptide) have been used for the mapping of epitopes of given antigens and will be discussed later. With these methods, it is now theoretically possible for staining of antigen-specific CD4+ T cells with tetramers to become a more useful technique.
Nonclassical tetramers
Defining the function and specificity of antigen-specific T cells is of obvious relevance to the analysis of vaccine-induced responses. While MHC class I tetramers are now in common use and class II tetramers are just starting to become widespread, some groups have explored alternative MHC tetramers to analyze T-cell function and specificity.
One such set of tetramers involves the CD1d protein. CD1 proteins constitute a distinct class of antigen-presenting molecules; they are distantly related to MHC molecules and are coded outside of the MHC locus. The nonpolymorphic CD1d glycoprotein associates with β2-microglobulin and binds glycolipids exemplified by α-GalCer Citation[30]. NKT cells, which possess TCRs specific for such molecules, represent up to 30% of the total lymphocyte population in the murine liver and are enriched to a lesser extent in the human liver Citation[31]. With the use of these tetramers, it is now possible to identify and study the distribution, frequency and modulation of NKT cells with standard tetramer techniques – this may be particularly relevant to innate responses induced by adjuvants Citation[32].
Applications of tetramer staining
General staining
A number of laboratories have developed standardized staining protocols for tetramers Citation[33], and we provide an outline here. Staining is most often performed at 37°C or at room temperature as this enhances TCR–tetramer interactions compared with staining on ice Citation[4]. Monoclonal antibodies may be used in concert for intracellular and surface staining, allowing abundant possibilities for analysis: typically tetramer staining is performed first to identify the population, especially before permeabilization and also before staining for CD8. Owing to the often low number and frequency of tetramer-positive cells, the use of positive, negative and isotope controls is advisable wherever possible. The use of whole blood should be considered as it often has an advantage for both clarity of staining and interpretation of results and only small amounts of blood (50 µl) Citation[34] are required. As the cells are untouched, there is low background and cleaner separation of the stained population of T cells for cell surface markers and tetramers. Such approaches also allow high-throughput analyses in a 96-well plate format Citation[34].
Tracking antigen-specific T-cell populations
The most obvious use of MHC–peptide tetramers in vaccinology is simply to track frequencies of CD8+ or, more rarely, CD4+ T cells in the circulation over time. This is readily achieved using conventional flow cytometry as described earlier although standardization of such techniques between laboratories to obtain comparable results still requires some work (see later). Key issues to address in each study include optimization and definition of background and cut-off, and definition of a positive response. The use of flow cytometers with multiparameter detection has allowed gating strategies, which improve some of these issues; this includes elimination of doublets, exclusion of dead cells and gating out of cells with nonspecific binding capacity such as B cells and monocytes.
However, flow cytometry still imposes a detection limit, typically in the region of 0.02% CD8+ T cells for detection. This is hard to overcome by simply using a larger input of cell numbers, but may be improved substantially using magnetic bead-enrichment protocols Citation[29,35–38]. Typically, these involve careful positive selection of cells that stain positive with a phycoerythrin (PE)-labeled tetramer, using beads coated with antibodies specific to PE. If one knows the starting number of cells and the number of cells selected with the PE magnetic beads, it is possible to calculate the percentage of tetramer positive cell for very low cell frequencies. This may be of use in analysis of relatively low-frequency CD8+ T-cell responses but may be essential for ex vivo detection of CD4+ T-cell responses. The technique retains good quantitation of the T-cell frequency in the critical 0.001–0.01% range.
Tetramer-guided epitope mapping
An advantage of the newly developed ‘empty’ tetramers is their use in identifying specific peptides from an antigen, a process described as tetramer-guided epitope mapping (TGEM) Citation[39–41]; this method is typically used for MHC class II and not class I molecules. Empty MHC molecules are loaded with peptide mixtures rather than an individual peptide. These peptide mixtures are overlapping peptides from an antigen of interest and are divided up into different pools, with each pool containing five to ten peptides. The pooled tetramers are used to analyze peripheral blood mononuclear cells that have been stimulated with the antigen of interest. Peptides from the positive pool are loaded onto the MHC molecule as individual peptides and are then analyzed. The specific tetramer that gives a positive staining provides a population of T cells of known MHC restriction and antigen specificity, and can be sorted by fluorescence-activated cell sorting (FACS), allowing phenotyping and genotyping of the cell.
Combined tetramer & functional assays
Proliferation
After infection or vaccination, there is an initial expansion of epitope-specific T cells; measuring this expansion and lifespan may be important in understanding the dynamics of T-cell responses and disease progression.
The proliferation of epitope-specific T cells can now be measured by combining standard techniques with tetramer staining. A widespread method utilizes carboxyfluorescein diacetate succinimidyl ester (CFSE; a nonspecific amine-reactive fluorescein derivative, which permeates membranes). Proliferative history is measured by serial halving of the dye during cell division for up to eight cycles and detected by flow cytometry Citation[42]. This technique in principle also allows the analysis of the precursor frequency from very rare cell populations (as in the enrichment techniques described previously) Citation[42,43]. Other techniques enable assessment of proliferation ex vivo. These employ measurement of intracellular markers that correlate to proliferation, such as DNA content (PI) and proteins expressed differentially in cycling and noncycling cells (KI67) Citation[44,45]. Typically, in these assays, cells will first be stained with tetramer then permeabilized and stained for the proliferation marker and/or surface markers, followed by FACS analysis.
Antiviral functions
Recently, ICS after peptide stimulation has proven to be a robust and sensitive technique for the analysis of T cells responding to viruses and vaccines. As for direct tetramer staining, the main limitation of the analysis of cells by ICS is cell frequency. However, with substantial responses (typically greater than 0.1%) it is possible to reliably analyze cytokines both singly and in combination. This technique can be combined with tetramer staining Citation[46], although technically this has proven to be problematic for a variety of reasons. First, after stimulation, there may be downregulation of TCRs, limiting the capacity to bind to tetramer Citation[47]. Second, if tetramer staining is performed before stimulation, there may be stimulation from the tetramer itself Citation[48], including induction of apoptotic pathways. These may be avoided, and it appears this may be somewhat variable from tetramer to tetramer, but it does impose limitations on the technique. In many cases, investigators perform in one analysis tetramer staining combined with phenotyping (see later) on unstimulated cells ex vivo and, in parallel, an analysis of the cytokine secretion profile by ICS. Although this does not provide analysis of the function of tetramer-binding cells on a per-cell basis, it does provide population-level information. Similarly, while it is not possible to analyze the cytolytic capacity of tetramer-positive populations in the same assay as their detection, analysis of upregulation of CD107A (LAMP1) as a marker of degranulation has been used as a surrogate by many investigators Citation[49].
A method used in analyzing T cells is the cytokine capture assay Citation[50]; this can be combined with tetramers Citation[51], allowing the recovery and purification of T cells based on their cytokine secretion and epitope specificity. To perform this assay, the cells are incubated with the tetramer of choice and then stimulated with the same epitope that the tetramer is constructed with, after which the cells are labeled with the cytokine catch antibody Citation[52]. This antibody consists of two biotin-labeled antibodies linked by an avidin molecule. One antibody is directed to the T-cell surface molecule CD45 and coupled to an antibody against the cytokine of interest. The cells are then incubated to allow secretion and capture of the cytokine, after which they will be labeled with a fluorochrome conjugated antibody directed against the cytokine of interest, and analyzed by FACS or sorted depending on their secretion profile. This method may significantly facilitate phenotypic and functional studies.
Phenotypic analyses
A wealth of data has been generated on the surface and intracellular expression of a range of molecular markers on antigen-specific T cells using tetramers, much of which is beyond the scope of this article. A fundamental issue in the development of effective vaccines is the quality of the T-cell response. There is no single specific molecular signature that predicts this, but combinations of markers may give a clue as to the overall nature of the response in terms of its recent proliferative history, its capacity for tissue homing and its state of activation. One widely used scheme is to divide tetramer-positive cells into three categories of central memory (Tcm), effector memory (Tem) and effector memory cells with re-expression of the RA isoform of CD45 (Temra) Citation[34,45,53,54]. These phenotypic markers overlay roughly with different functional and proliferative capacities, and have been correlated with different outcomes in different infections. However, none are currently a surrogate marker for a protective response in vaccine studies. Similarly, new data from the lymphocytic choriomeningitis model and translated into human chronic viral infection have illustrated some of the molecular pathways associated with downregulation of the T-cell responses and the induction of exhaustion Citation[55]. This includes the expression of PD-1 Citation[56], as well as other inhibitory molecules such as cytotoxic T-lymphocyte antigen 4 Citation[57]. However, since many of these pathways are also upregulated on activated cells, it is not possible to accurately define the function of the cell or its in vivo protective capacity from such phenotyping. Phenotypic analyses therefore remain an important research tool and currently provide important correlative data in new vaccine studies, rather than providing an end point.
In situ tetramer staining
Until recently, tetramers have been limited to ex vivo studies but they can be used to visualize antigen-specific CD8+ T cells in tissue with their spatial relationship to other intact cells in so-called in situ tetramer (IST) staining Citation[58–60]. Fresh unfixed tissue is ideally used to allow for flexibility in the TCR for binding to the tetramer. A vibratome (TPI) may be used to generate 20-µm thick fresh tissue sections, which are then stained with tetramer and cell surface markers overnight. Both direct and indirect IST staining have been used in staining these sections that are then fixed. This method has only been used successfully by a few groups, and this has mainly been in murine models Citation[58], although some human studies have been performed Citation[60].
Adoptive T-cell transfer
Tetramers may be used for the selective isolation of functionally active antigen-specific T cells – this could be regarded therefore as tetramer-guided ‘passive’ immunization. A specific cell population can be identified using tetramers and then enriched, expanded in culture and transferred at high frequency. To date, there is some experience of such techniques in man in the treatment of cytomegalovirus infections and HIV, and it has been used with or without expansion of the tetramer-positive cells Citation[61–63], However, in the future, such programs demand the development of Good Manufacturing Practice-quality tetramers.
Sorting antigen-specific T cells
A method increasingly utilized is tetramers in combination with cell-sorting techniques for clonotypic expression. Stained cells can be sorted by FACS or by the use of magnetic anti-fluorochrome microbeads, producing pure populations of epitope-specific T cells of high purity Citation[64]. The live unfixed cells can then be analyzed functionally or their RNA can be extracted for expression analysis. More detailed analysis of the cellular RNA content will, in the future, provide an unprecedented wealth of information on the phenotype and function of epitope-specific T cells at the single cell level Citation[64–66].
Modified tetramers
Recently, mutant tetramers have been constructed that possess modified MHC class I backbones. These mutations either diminish or increase CD8/MHC binding interactions, and thus provide a way of sampling different repertoires of T cells with respectively higher or lower TCR avidities Citation[67,68]. The use of such techniques has recently been reviewed elsewhere, and they may be of value in the future in defining responses of sufficient avidity (as indicated by CD8 independence) to provide information about efficacy.
Combinatorial tetramer staining
This very new technique may be of real value in simultaneous monitoring of diverse responses in a single sample. The ability to analyze only single specificities has long been a major limitation of tetramer-based approaches. Davis’s group, who generated the first tetramers Citation[1], have published a method whereby different tetramers can be mixed using distinct fluorochromes, such that each tetramer is represented by some but not all fluorochromes Citation[69]. Thus, using a limited number of colors (e.g., four), 15 different specificities might be detected simultaneously in the sample . This increases exponentially with color, so that with five colors this theoretically rises to 31 and with six to 63. In combination with the recent advances in multiplexing tetramer construction outlined earlier, this approach could greatly enhance the use of tetramers to track broader specificities of induced T-cell responses.
Conclusion & future directions
MHC tetramers can only detect a subset of the total pool of antigen-specific T cells directed against specific infectious agents. The main limitations of this technology lie in the fact that it requires cloned and characterized MHC molecules, MHC-matched tissue and knowledge of the specific peptide sequence. Through enormous effort from the immunologic community, such reagents and information are in fact available for a large number of major vaccine targets, and continue to expand over time. However, detailed tetramer-based analysis still typically provides only a snapshot of the target cell population and other methods using pooled peptides and/or recombinant antigens and vectors are necessary to provide more complete analyses of breadth in most cases. The ability to provide multiplexed sets of peptide-loaded tetramers and to analyze them simultaneously should provide an important improvement in the technology to circumvent some aspects of this problem. However, tetramer analysis will always need to be combined with functional analyses and also with techniques that screen broadly for responses independent of MHC type, especially in complex or variable viruses.
For vaccinologists, the advantages of using tetramers are their specificity and sensitivity and the capacity for this technology to be carried out in conjunction with other assays. Surface and intracellular staining can be carried out, allowing phenotyping of epitope-specific cells, and recent advances have led to functional assays being combined with tetramer staining and tetramer-positive cells being sorted and genotyped or cloned. For a given vaccine study, the advantages of tetramers may be limited by the disadvantage that only a small fraction of the response can be measured. This may be partially overcome in the future using a multiplexed approach, but will remain a limitation
The future of such assays demands some further standardization between laboratories. This is an issue that affects most assays involving T-cell immunology and is not restricted to tetramers. Indeed, tetramer staining circumvents some of the issues involved in many such assays since they do not rely on functional readouts. However, it is important as the field evolves to develop systems to allow better comparisons between trials and experiments. Such a proposal has been recently put forward by Janetzki and colleagues Citation[70] and is described as minimal information about T cell assays (MIATA; details available at Citation[101]). Some investigators Citation[33] have proposed the development of core facilities to drive such improvements in human immunomonitoring, some of which are emerging (e.g., Citation[102–104]). The use of web-based protocols and the development of such centers should serve to accelerate this movement.
In conclusion, MHC–peptide tetramers have just passed their 13th birthday and there are many reasons to celebrate. As the technology matures and develops, new opportunities will arise to use the technology in different settings and in combination with different assays. At the same time, as the field matures and tetramers move from research tools towards clinical use, some consensus needs to be reached to allow the very best aspects to be utilized and the obvious limitations to be avoided. Thus, many future improvements in the clinical setting may result not so much from further new technology itself, but rather from more strategic and harmonized use of the technology available. The field of vaccinology has benefited hugely from these reagents and, driven by the demand for new T-cell based vaccines and immunotherapy, it will continue to do so.
Expert commentary
MHC–peptide tetramers are now robust tools for immunomonitoring. They can provide accurate quantification of antigen-specific T cells after vaccination, although they should be combined with methods to assess the breadth of immune responses as well as their functionality in order to obtain a full picture. Laboratories using these tools should try to develop or join consortia in order to maintain standardization across sites.
Five-year view
Tetramers will continue to be developed to provide a wider base of HLA molecules and enable them to be combined better with functional assays. The use of combinational approaches both for production of tetramers and for flow cytometric analysis should allow in-depth characterization of novel vaccines as well as providing breadth and functional data.
Tetramers are widely used and will become a standard laboratory technique with the current improvements in combinational approaches and an increase in the number of HLA molecules available. They currently provide an in-depth analysis of specific T cells and, with these new methods, should provide a wealth of unprecedented information on specific T cells.
Box 1. Current species availability of MHC class I and II tetramers.
MHC class I
• Humans
• Mice
• Chimpanzees
• Chickens
• Macaques
• Sooty mangabeys
• Swine
MHC class II
• Humans
• Mice
• Chimpanzees
• Chickens
Key issues
• Tetramer construction: in this article, we have summarized the improvements in the construction of tetramers, and the streamlining of the process so that it is less labor intensive.
• Combined tetramer and functional assay: using assays in conjunction with tetramers such as cytokine capture we are starting to gain a greater insight into the specific function of epitope-specific cells.
• Tetramer-guided epitope mapping: this method will allow elucidation of previously unknown epitope-specific cells by using tetramers loaded with peptide pools.
• Sorting tetramer-positive cells: these cells can then be studied in greater depth either by gene array or they can be used in killing assays, which should provide the next great wealth of information on T cells.
• Adoptive immunotherapy: using tetramers to select functionally active epitope-specific cells, then expanding and transferring these cells to patients, may provide new treatments for diseases.
Financial & competing interests disclosure
This work was supported by the Wellcome Trust, the MRC, the James Martin School for the 21st Century Oxford, the NHS U19A1082830 and the NIHR Biomedical Research Centre Programme Oxford. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- Altman JD, Moss PA, Goulder PJ et al. Phenotypic analysis of antigen-specific T lymphocytes. Science274(5284), 94–96 (1996).
- Valitutti S, Muller S, Cella M, Padovan E, Lanzavecchia A. Serial triggering of many T-cell receptors by a few peptide–MHC complexes. Nature375(6527), 148–151 (1995).
- Laugel B, Boulter JM, Lissin N et al. Design of soluble recombinant T cell receptors for antigen targeting and T cell inhibition. J. Biol. Chem.280(3), 1882–1892 (2005).
- Whelan JA, Dunbar PR, Price DA et al. Specificity of CTL interactions with peptide–MHC class I tetrameric complexes is temperature dependent. J. Immunol.163(8), 4342–4348 (1999).
- Cameron TO, Norris PJ, Patel A et al. Labeling antigen-specific CD4+ T cells with class II MHC oligomers. J. Immunol. Methods268(1), 51–69 (2002).
- Kuroda MJ, Schmitz JE, Lekutis C et al. Human immunodeficiency virus type 1 envelope epitope-specific CD4+ T lymphocytes in simian/human immunodeficiency virus-infected and vaccinated rhesus monkeys detected using a peptide–major histocompatibility complex class II tetramer. J. Virol.74(18), 8751–8756 (2000).
- Mallet-Designe VI, Stratmann T, Homann D, Carbone F, Oldstone MB, Teyton L. Detection of low-avidity CD4+ T cells using recombinant artificial APC: following the antiovalbumin immune response. J. Immunol.170(1), 123–131 (2003).
- Stratmann T, Martin-Orozco N, Mallet-Designe V et al. Susceptible MHC alleles, not background genes, select an autoimmune T cell reactivity. J. Clin. Invest.112(6), 902–914 (2003).
- Liu G, Wang Q, Tong T et al. Construction and functional test of a chicken MHC-I (BF2*15)/peptide tetramer. Vet. Immunol. Immunopathol.122(1–2), 1–7 (2008).
- Mealey RH, Sharif A, Ellis SA, Littke MH, Leib SR, McGuire TC. Early detection of dominant Env-specific and subdominant Gag-specific CD8+ lymphocytes in equine infectious anemia virus-infected horses using major histocompatibility complex class I/peptide tetrameric complexes. Virology339(1), 110–126 (2005).
- Oleksiewicz MB, Kristensen B, Ladekjaer-Mikkelsen AS, Nielsen J. Development of a rapid in vitro protein refolding assay which discriminates between peptide-bound and peptide-free forms of recombinant porcine major histocompatibility class I complex (SLA-I). Vet. Immunol. Immunopathol.86(1–2), 55–77 (2002).
- Donahoe SM, Moretto WJ, Samuel RV et al. Direct measurement of CD8+ T cell responses in macaques infected with simian immunodeficiency virus. Virology272(2), 347–356 (2000).
- Dunbar PR, Ogg GS, Chen J, Rust N, van der Bruggen P, Cerundolo V. Direct isolation, phenotyping and cloning of low-frequency antigen-specific cytotoxic T lymphocytes from peripheral blood. Curr. Biol.8(7), 413–416 (1998).
- Rout N, Else JG, Yue S, Connole M, Exley MA, Kaur A. Paucity of CD4+ natural killer T (NKT) lymphocytes in sooty mangabeys is associated with lack of NKT cell depletion after SIV infection. PLoS One5(3), e9787 (2010).
- Niemiec PK, Read LR, Sharif S. Synthesis of chicken major histocompatibility complex class II oligomers using a baculovirus expression system. Protein Expr. Purif.46(2), 390–400 (2006).
- Garboczi DN, Hung DT, Wiley DC. HLA-A2–peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl Acad. Sci. USA89(8), 3429–3433 (1992).
- Leisner C, Loeth N, Lamberth K et al. One-pot, mix-and-read peptide–MHC tetramers. PLoS One3(2), e1678 (2008).
- Batard P, Peterson DA, Devêvre E et al. Dextramers: new generation of fluorescent MHC class I/peptide multimers for visualization of antigen-specific CD8+ T cells. J. Immunol. Methods310(1–2), 136–148 (2006).
- Christmas SE, Halliday D, Lawton N et al. Cytomegalovirus-specific CD8+ T cells do not develop in all renal transplant patients at risk of virus infection. Transplant Immunol.22(1–2), 99–104 (2009).
- Bakker AH, Hoppes R, Linnemann C et al. Conditional MHC class I ligands and peptide exchange technology for the human MHC gene products HLA-A1, -A3, -A11, and -B7. Proc. Natl Acad. Sci. USA105(10), 3825–3830 (2008).
- Soen Y, Chen DS, Kraft DL, Davis MM, Brown PO. Detection and characterization of cellular immune responses using peptide–MHC microarrays. PLoS Biol.1(3), E65 (2003).
- Crawford F, Kozono H, White J, Marrack P, Kappler J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity8(6), 675–682 (1998).
- Yang J, Jaramillo A, Shi R, Kwok WW, Mohanakumar T. In vivo biotinylation of the major histocompatibility complex (MHC) class II/peptide complex by coexpression of BirA enzyme for the generation of MHC class II/tetramers. Hum. Immunol.65(7), 692–699 (2004).
- Kozono H, White J, Clements J, Marrack P, Kappler J. Production of soluble MHC class II proteins with covalently bound single peptides. Nature369(6476), 151–154 (1994).
- Malherbe L, Filippi C, Julia V et al. Selective activation and expansion of high-affinity CD4+ T cells in resistant mice upon infection with Leishmania major.Immunity13(6), 771–782 (2000).
- Novak EJ, Liu AW, Nepom GT, Kwok WW. MHC class II tetramers identify peptide-specific human CD4+ T cells proliferating in response to influenza A antigen. J. Clin. Invest.104(12), R63–R67 (1999).
- Moro M, Cecconi V, Martinoli C et al. Generation of functional HLA-DR*1101 tetramers receptive for loading with pathogen- or tumour-derived synthetic peptides. BMC Immunol.6, 24 (2005).
- Lebowitz MS, O’Herrin SM, Hamad AR et al. Soluble, high-affinity dimers of T-cell receptors and class II major histocompatibility complexes: biochemical probes for analysis and modulation of immune responses. Cell. Immunol.192(2), 175–184 (1999).
- Day CL, Seth NP, Lucas M et al.Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J. Clin. Invest.112(6), 831–842 (2003).
- Porcelli SA. The CD1 family: a third lineage of antigen-presenting molecules. Adv. Immunol.59, 1–98 (1995).
- Benlagha K, Bendelac A. CD1d-restricted mouse V α 14 and human V α 24 T cells: lymphocytes of innate immunity. Semin. Immunol.12(6), 537–542 (2000).
- Matsuda JL, Naidenko OV, Gapin L et al. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J. Exp. Med.192(5), 741–754 (2000).
- Guillaume P, Dojcinovic D, Luescher IF. Soluble MHC–peptide complexes: tools for the monitoring of T cell responses in clinical trials and basic research. Cancer Immun.9, 7 (2009).
- Komatsu H, Sierro S, V Cuero A, Klenerman P. Population analysis of antiviral T cell responses using MHC class I–peptide tetramers. Clin. Exp. Immunol.134(1), 9–12 (2003).
- Scriba TJ, Zhang HT, Brown HL et al. HIV-1-specific CD4+ T lymphocyte turnover and activation increase upon viral rebound. J. Clin. Invest.115(2), 443–450 (2005).
- Day CL, Kiepiela P, Leslie AJ et al. Proliferative capacity of epitope-specific CD8 T-cell responses is inversely related to viral load in chronic human immunodeficiency virus type 1 infection. J. Virol.81(1), 434–438 (2007).
- Barnes E, Ward SM, Kasprowicz VO, Dusheiko G, Klenerman P, Lucas M. Ultra-sensitive class I tetramer analysis reveals previously undetectable populations of antiviral CD8+ T cells. Eur. J. Immunol.34(6), 1570–1577 (2004).
- Lucas M, Day CL, Wyer JR et al.Ex vivo phenotype and frequency of influenza virus-specific CD4 memory T cells. J. Virol.78(13), 7284–7287 (2004).
- Novak EJ, Liu AW, Gebe JA et al. Tetramer-guided epitope mapping: rapid identification and characterization of immunodominant CD4+ T cell epitopes from complex antigens. J. Immunol.166(11), 6665–6670 (2001).
- Reijonen H, Kwok WW. Use of HLA class II tetramers in tracking antigen-specific T cells and mapping T-cell epitopes. Methods29(3), 282–288 (2003).
- Kwok WW, Gebe JA, Liu A et al. Rapid epitope identification from complex class-II-restricted T-cell antigens. Trends Immunol.22(11), 583–588 (2001).
- Lyons AB. Analysing cell division in vivo and in vitro using flow cytometric measurement of CFSE dye dilution. J. Immunol. Methods243(1–2), 147–154 (2000).
- Takata H, Takiguchi M. Three memory subsets of human CD8+ T cells differently expressing three cytolytic effector molecules. J. Immunol.177(7), 4330–4340 (2006).
- Oxenius A, Gunthard HF, Hirschel B et al. Direct ex vivo analysis reveals distinct phenotypic patterns of HIV-specific CD8+ T lymphocyte activation in response to therapeutic manipulation of virus load. Eur. J. Immunol.31(4), 1115–1121 (2001).
- Sierro S, Rothkopf R, Klenerman P. Evolution of diverse antiviral CD8+ T cell populations after murine cytomegalovirus infection. Eur. J. Immunol.35(4), 1113–1123 (2005).
- Appay V, Nixon DF, Donahoe SM et al. HIV-specific CD8+ T cells produce antiviral cytokines but are impaired in cytolytic function. J. Exp. Med.192(1), 63–75 (2000).
- Holmberg K, Mariathasan S, Ohteki T, Ohashi PS, Gascoigne NR. TCR binding kinetics measured with MHC class I tetramers reveal a positive selecting peptide with relatively high affinity for TCR. J. Immunol.171(5), 2427–2434 (2003).
- Delon J, Gregoire C, Malissen B et al. CD8 expression allows T cell signaling by monomeric peptide–MHC complexes. Immunity9(4), 467–473 (1998).
- Betts MR, Brenchley JM, Price DA et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J. Immunol. Methods281(1–2), 65–78 (2003).
- Streeck H, Cohen KW, Jolin JS et al. Rapid ex vivo isolation and long-term culture of human Th17 cells. J. Immunol. Methods333(1–2), 115–125 (2008).
- Van Overtvelt L, Wambre E, Maillere B et al. Assessment of Bet v 1-specific CD4+ T-cell responses in allergic and nonallergic individuals using MHC class II peptide tetramers. J. Immunol.180(7), 4514–4522 (2008).
- Assenmacher M, Lohning M, Radbruch A. Detection and isolation of cytokine secreting cells using the cytometric cytokine secretion assay. Curr. Protoc. Immunol. Chapter 6, Unit 6 27 (2002).
- Snyder CM, Cho KS, Bonnett EL, van Dommelen S, Shellam GR, Hill AB. Memory inflation during chronic viral infection is maintained by continuous production of short-lived, functional T cells. Immunity29(4), 650–659 (2008).
- Karrer U, Sierro S, Wagner M et al. Memory inflation: continuous accumulation of antiviral CD8+ T cells over time. J. Immunol.170(4), 2022–2029 (2003).
- Shin H, Blackburn SD, Intlekofer AM et al. A role for the transcriptional repressor Blimp-1 in CD8+ T cell exhaustion during chronic viral infection. Immunity31(2), 309–320 (2009).
- Wang W, Lau R, Yu D, Zhu W, Korman A, Weber J. PD1 blockade reverses the suppression of melanoma antigen-specific CTL by CD4+ CD25Hi regulatory T cells. Int. Immunol.21(9), 1065–1077 (2009).
- Jain N, Nguyen H, Chambers C, Kang J. Dual function of CTLA-4 in regulatory T cells and conventional T cells to prevent multiorgan autoimmunity. Proc. Natl Acad. Sci. USA107(4), 1524–1528 (2010).
- Skinner PJ, Daniels MA, Schmidt CS, Jameson SC, Haase AT. Cutting edge: in situ tetramer staining of antigen-specific T cells in tissues. J. Immunol.165(2), 613–617 (2000).
- Li Q, Skinner PJ, Ha SJ et al. Visualizing antigen-specific and infected cells in situ predicts outcomes in early viral infection. Science323(5922), 1726–1729 (2009).
- De Vries IJ, Bernsen MR, van Geloof WL et al.In situ detection of antigen-specific T cells in cryo-sections using MHC class I tetramers after dendritic cell vaccination of melanoma patients. Cancer Immunol. Immunother.56(10), 1667–1676 (2007).
- Casalegno-Garduno R, Schmitt A, Yao J et al. Multimer technologies for detection and adoptive transfer of antigen-specific T cells. Cancer Immunol. Immunother.59(2), 195–202 (2010).
- Savage P, Millrain M, Dimakou S, Stebbing J, Dyson J. Expansion of CD8+ cytotoxic T cells in vitro and in vivo using MHC class I tetramers. Tumour Biol.28(2), 70–76 (2007).
- Cobbold M, Khan N, Pourgheysari B et al. Adoptive transfer of cytomegalovirus-specific CTL to stem cell transplant patients after selection by HLA–peptide tetramers. J. Exp. Med.202(3), 379–386 (2005).
- Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med.205(3), 625–640 (2008).
- Wherry EJ, Ha SJ, Kaech SM et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity27(4), 670–684 (2007).
- Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory CD8 T cell differentiation. Cell111(6), 837–851 (2002).
- Melenhorst JJ, Scheinberg P, Chattopadhyay PK et al. Detection of low avidity CD8+ T cell populations with coreceptor-enhanced peptide–major histocompatibility complex class I tetramers. J. Immunol. Methods338(1–2), 31–39 (2008).
- Wooldridge L, Lissina A, Vernazza J et al. Enhanced immunogenicity of CTL antigens through mutation of the CD8 binding MHC class I invariant region. Eur. J. Immunol.37(5), 1323–1333 (2007).
- Newell EW, Klein LO, Yu W, Davis MM. Simultaneous detection of many T-cell specificities using combinatorial tetramer staining. Nat. Methods6(7), 497–499 (2009).
- Janetzki S, Britten CM, Kalos M et al. ‘MIATA’ – minimal information about T cell assays. Immunity31(4), 527–528 (2009).
Websites
- MIATA Reporting Framework www.miataproject.org
- Cancer Immunity www.cancerimmunity.org
- Stanford School of Medicine. HIMC – Human Immune Monitoring Center http://iti.stanford.edu/research/human_immune_monitoring.html
- Oxford Biomedical Research Centre: Translation Immunology Laboratory http://oxfordbrc.org/immunity/research-areas/172
