Abstract
Purpose
The diagnosis of irritable bowel syndrome (IBS) is based on symptom-based criteria due to lack of reliable disease-specific biomarkers. Gut microbiota is perturbed in IBS and when comparing different methods used to analyze gut microbiota, the results might be obscured. Therefore, in this systematic review we aimed to investigate the profile of fecal bacterial markers and dysbiosis index (DI) in patients with IBS and IBS subgroups compared to healthy controls (HCs) conducted by the same method (GA-map Dysbiosis Test based on16S rRNA sequencing).
Material and Method
We searched PubMed, EMBASE (Ovid) and Cochrane Library for case-control studies comparing fecal gut microbiota analyzed with the GA-map® Dysbiosis Test (Oslo, Norway) in patients with IBS and HCs. Our outcomes were the difference in fecal bacterial markers and DI in patients with IBS and IBS subgroups compared to HCs.
Results
The search identified 28 citations; five articles were included. Most studies evaluated fecal bacterial markers and DI in patients with diarrhea-predominant IBS (IBS-D). Results of fecal bacteria profile in IBS and IBS subgroups compared to HCs are inconsistent, however, two studies showed increased levels of Ruminococcus gnavus in IBS-D compared to HCs and results of DI indicated IBS and IBS subgroups (especially IBS-D) having higher DI compared to HCs.
Conclusion
This systematic review revealed inconsistent findings in respect to differences in bacterial markers between IBS and IBS subgroups with HCs in studies using the GA-map Dysbiosis Test based on 16S rRNA sequencing. However, the test is quite novel, and few studies have used the method so far. More research comparing fecal microbiota profile differences in IBS and IBS subgroups compared to HCs utilizing the same method of analysis is needed to give us further insight into the gut bacteria profile in IBS and the clinical consequences of intestinal dysbiosis.
Introduction
Irritable bowel syndrome (IBS) is a disorder of the brain-gut axis interactionCitation1 affecting 4–10% of the general population.Citation2 Today, the diagnosis is based on exclusion of organic diseases and fulfilling the Rome IV criteriaCitation1 which is based on symptoms of recurrent abdominal pain at least one day per week associated with stool irregularities.Citation3 IBS is divided in four subtypes based on the predominant stool consistency: IBS with predominantly diarrhea (IBS-D), with predominantly constipation (IBS-C), with a mix of diarrhea and constipation (IBS-M) and unclassified subtype (IBS-U).Citation3 The symptoms are experienced from mild to severe and often comes with somatic and psychiatric comorbidities such as fibromyalgia, fatigue, depression and anxiety, and impacts the quality of life in these patients.Citation4–6 It also gives a profound burden on the healthcare system and the society.Citation6
The etiology of IBS is complex and multifactorial, including altered brain-gut-microbiota axis, visceral hypersensitivity, low-grade inflammation, intestinal dysbiosis, and impaired gut barrier functions and integrity.Citation1,Citation7–9 Dysbiosis is an deviation from normobiosis and is defined as an imbalance in bacterial composition.Citation4 Today, the manipulation of gut microbiota is suggested to be a target for therapy of IBSCitation7 but the gut microbiota profile in IBS is still not fully understood as the literature reports inconsistent results. Several different methods are available and used for studying the gut microbiota and the costs are becoming more affordable.Citation7 However, when comparing the fecal microbiota profile across different methods, the results might be obscured.
Therefore, in this systematic review we aimed to investigate the profile of fecal bacterial markers and dysbiosis index (DI) in patients with IBS and IBS subgroups compared to healthy controls (HCs) conducted by the same method of analysis, GA-map Dysbiosis Test.Citation10 The test is based on 16S rRNA sequencing, is commercially available and is an easy to use method which aims to identify and characterize dysbiosis by determining deviation from normobiosis in fecal samples in IBS and IBD. Hopefully, this will give us further insight into the gut bacteria profile in IBS and the clinical consequences of intestinal dysbiosis.
Materials and Methods
Search Strategy
We performed a systematic review with relevant literature retrieved from the databases PubMed, EMBASE (Ovid) and Cochrane Library to identify case-control studies comparing fecal gut microbiota analyzed with the GA-map Dysbiosis Test in patients with IBS and HCs. The search terms used to identify potentially related publications included «Genetic analysis AND Dysbiosis test AND Irritable bowel syndrome», with results by year 2015 to January 2023. Two authors independently reviewed the studies retrieved by the search strategy and excluded trials based on titles, abstracts, and full-text analysis. We followed the checklist and flowchart of the Preferred Reporting of Systematic Reviews and Meta-Analyses (PRISMA) for this systematic review.
Study Selection
The inclusion criteria () were studies investigating fecal microbiota in adult IBS patients compared with HCs, IBS patients classified according to the ROME II–IV criteria, GA-map Dysbiosis Test to analyze fecal bacterial samples, samples from stool, written in English, published from 2015 to January 2023 and articles published as a full article. The exclusion criteria were literature published before 2015, not written in English, fecal microbiota not analyzed with the GA-map Dysbiosis Test or not case-control studies.
Table 1 Inclusion Criteria for Articles Included in the Systematic Review
Choice of Outcome
The primary outcome was differences in fecal bacterial markers and DI in patients with IBS compared to HCs. The secondary outcome was differences in bacterial markers and DI in subgroups of IBS compared to HCs. We classified the primary and secondary outcomes into three categories; significantly increased in IBS patients, significantly decreased in IBS patients and no significant (NS) difference between IBS patients and HCs. The DI was estimated as mean or median.
Fecal Bacteria and Dysbiosis Analysis
GA-map Dysbiosis Test (GA-map® Dysbiosis Test, Oslo, Norway)Citation10 was developed to identify and characterize dysbiosis by determining deviation from normobiosis in fecal samples in patients with IBS and IBD. It gives an algorithmically Dysbiosis Index (DI) from 1 to 5 where a DI value of 2 or lower being non-dysbiotic and 3–5 being dysbiotic. The higher DI above 2, the more it is considered to deviate from normobiosis and being dysbiotic. Transient luminal bacteria with no diagnostic values have the potential to obscure diagnostic results. Therefore, the probe set was based on intestinal microbiota observed in IBS and IBD in the literature and each probe was selected based on the ability to distinguish between IBS and IBD and HCs.Citation10 The technology uses 54 bacterial markers, based on the 16S rRNA sequences in seven variable regions (V3-V9) and measures relative abundance of bacteria according to strength of fluorescent signal detection. This provides a unique opportunity to investigate changes in gut microbiota profiles which potentially might be associated with GI-related disorders.
Eligibility Assessment and Data Extraction
To reduce the reporting error and bias in data manipulation, both authors independently reviewed the selected studies for complete analysis. One study author extracted the data and entered it into a spreadsheet, while the other study author evaluated the process. When there was a discrepancy between reviewers, we re-checked and discussed the data together to reach an agreement by consensus. The data included are presented in ; distribution of age and sex, country in which the research was conducted, the size of the IBS and HC groups, IBS subtypes, details of intervention, multiple comparison correction, dysbiosis index and bacterial markers difference with a significant P value < 0.05.
Table 2 Characteristics of Included Studies in the Systematic Review
Quality Assessment
We applied the Newcastle-Ottawa Scale (NOS) for assessing the quality of included case-control studies in this review (data presented in ). The Newcastle-Ottawa Scale consists of 3 domains (maximum nine stars); selection (is the case definition adequate, representativeness of the cases, selection of controls, definition of controls); comparability (comparability of baseline characteristics); and exposure (ascertainment of exposure, same method of ascertainment for cases and controls, attrition rate). We decided that sex and age was the most important baseline characteristics to be described and compared in both groups.
Table 3 Quality of Each Study Included by Newcastle Ottawa Scale (NOS)
Results
Study Selection and Characteristics
Overall, 28 citations were retrieved (): six citations were duplicates and four citations were rejected based on their title. Within the 22 remaining studies, 13 were excluded based on the abstract or full-text review as nine studies had no healthy controls, two studies were conference abstracts and two papers included repetition of data. In total, five studies investigating fecal microbiota composition and/or DI were included in the systematic review.
Figure 1 PRISMA flow diagram for depicting the literature search in PubMed (Medline) for this systematic review.
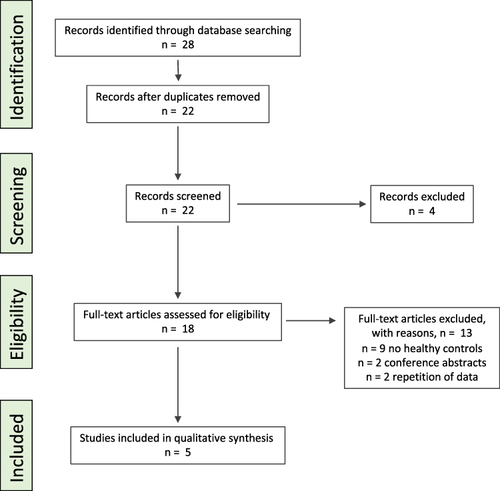
Study populations across the studies were conducted from Sweden,Citation10,Citation11,Citation13 Norway,Citation10,Citation14 Denmark,Citation10 SpainCitation10 and Germany.Citation12 All studies had case-control design, were published in the period of 2015 to January 2023 and fecal microbiota was assessed with the GA-map Dysbiosis Test in adults of both genders.
The Newcastle Ottawa Scale () showed all relevant studies provided an adequate explanation in the definition and selection method for IBS patients and HCs with IBS classified according to Rome II–IV criteria by a physician, and HCs did not have any organic disease or gastrointestinal (GI) symptoms. All studies demonstrated comparable data of both sex and age in IBS and HCs, hence, one study had significantly higher median age in IBS compared to HCsCitation11 and as expected, all studies had an overrepresentation of females in both groups except from Ahluwalia et al 2021Citation11 having the same number for males and females in HCs, and Mazzawi et al 2018Citation14 having more males than females in IBS (). Two out of five articles reported multiple comparison correctionCitation12,Citation14 and all studies used a significant p-value < 0.05 for bacterial marker and/or DI difference.
In terms of IBS subtype, three out of five studies included both IBS-D and IBS-CCitation10–12 where two also included IBS-M.Citation10,Citation11 The two remaining articles included only IBS-D.Citation13,Citation14 One study did not investigate fecal microbiota composition between IBS and HCs nor across IBS subgroups but investigated DI.Citation10 One study investigated the DI for IBS,Citation10 three studies investigated the DI for IBS-D,Citation10,Citation12,Citation14 two studies investigated DI for IBS-CCitation10,Citation12 and only one study investigated DI for IBS-M.Citation10 In respect to bacterial markers, one study investigated IBS compared to HCs,Citation11 three studies investigated IBS-DCitation12–14 one study investigated IBS-CCitation12 and no studies investigated the difference in bacterial markers in IBS-M compared to HCs.
Fecal Microbiota Composition
Findings for fecal microbiota are summarized in . Among five studies included in this review, the reported main findings are overall inconsistent. For differences between IBS and HCs, one study reported enrichment of Acinetobacter junii and Akkermansia muciniphila and decreased levels of Clostridium sp. in IBS compared to HCs. For differences between IBS subgroups and HCs only IBS-D was reported to have significant findings; two studies reported increased levels of Ruminococcus gnavus, one study reported enrichment of Proteobacteria and Dorea spp. and one study reported decreased levels of Bacteroides pectinophilus, Actinobacteria and Bifidobacteria compared to HCs.
Table 4 Differences in Bacterial Markers Between IBS and IBS Subgroups and Healthy Controls (HCs)
Dysbiosis Index
Findings for DI are summarized in . The reported main findings indicate that IBS and IBS subgroups have higher levels of DI compared to HCs. For differences between IBS and HCs, one study found a significant increase in DI in IBS compared to HCs. For differences between IBS subgroups and HCs: two studies found IBS-D with increasedCitation10,Citation14 and one study with a non-significant increasedCitation12 DI compared to HCs. In IBS-C one study found a significant and one study found a non-significant increase in DI compared to HCs. For the only study investigating IBS-M, the DI was significantly increased in comparison to HCs.
Table 5 Dysbiosis Index in IBS and IBS Subgroups Compared to Healthy Controls (HCs)
Discussion
To our knowledge, this is the first systematic review investigating differences in fecal bacterial profile and DI between IBS and IBS subgroups compared to HCs performed by using GA-map Dysbiosis Test based on the 16s rRNA gene of bacterial species or groups. Based on only five studies included the results was inconsistent. However, two studies showed an increase in Ruminococcus gnavus in IBS-D compared to HCs and results of DI indicated IBS and IBS subgroups (especially IBS-D) having higher DI compared to HCs.
Today, there are many methods used to investigate the gut microbiota profile in patients with IBS, and recently we have been able to increase the exploration due to progress in microbial analytic techniques.Citation4 Studies have shown that IBS have a different microbiota profile compared to HCs, but the existing literature is inconclusive.Citation15 There are many factors which makes it challenging to interpret the results of gut microbiota profile. Firstly, the gut microbiota is highly dynamic and is affected especially by diet, medications, ethnicity, and geographical deviations. Secondly, the samples can be taken at different sites such as in stool or mucosa, and thirdly, the results can be obscured when comparing findings across different methods.Citation15,Citation16 All these factors might contribute to the inconclusive findings of gut microbiota in IBS and is needed to be taken into consideration when interpreting results of gut microbiota.
In the present review, only one study investigated the difference in fecal bacterial markers between IBS and HCs: increased levels of Acinetobacter junii (phyla Proteobacteria) and Akkermansia muciniphila (phyla Verrucomicrobiota) and decreased levels of Clostridium sp. (phyla Firmicutes) in IBS compared to HCs, and only one study compared the DI between the two groups: DI was significantly higher in IBS compared to HCs.
The observations of differences in bacterial composition are not similar to previous findings conducted with several different methods of bacterial analysis. A systematic reviewCitation17 investigating differences in gut microbiota in IBS compared to HCs conducted with 16S rRNA-targeted sequencing observed consistent findings which included increased level of the phyla Firmicutes and reduced level of the phyla Bacteroidetes and increased Firmicutes/Bacteroidetes ratio, higher levels of Clostridia and Clostridiales (phyla Firmicutes), and lower levels of Bacteroidia and Bacteroidales (phylum Bacteroidetes) in IBS. The studies were discrepant in results of changes in alpha-diversity (specie richness), observing both higher, lower and no difference between IBS and HCs using the Shannon index and Chao1 index. However, almost similar to our findings, the authors found a trend of IBS having lower alfa-diversity in comparison to HCs. A systematic review conducted by Pittayanon et al 2019Citation15 based on stool and colonic tissue microbiota analyzed with different methods found that the difference between IBS and HCs included increased levels of Enterobacteriaceae (family, phylum Proteobacteria), Lactobacillaceae (family, phylum Firmicutes) and Bacteroides (genus, phyla Bacteroidetes), and reduced levels of uncultured Clostridiales I (phyla Firmicutes), genus Faecalibacterium (including Faecalibacterium prausnitzii, phyla Firmicutes) and genus Bifidobacterium (phyla Actinobacteria) in IBS compared to HCs. One study showed increased levels and another no significant difference in the abundance of Acinetobacter (phylum Proteobacteria) in IBS compared to HCs. The diversity of microbiota was either decreased or not different in IBS compared to HCs. The author addressed that there were no two studies reporting the same differences in OTUs.Citation15 Another systematic review and meta-analysis of fecal microbiota profile in IBS compared to HCs using quantitative real-time PCR analysis found a reduced abundance of Lactobacillus (phyla Firmicutes), Bifidobacterium (phyla Actinobacteria) and F. Prausnitzii (phyla Firmicutes) in IBS compared to HCs.Citation18
The abovementioned systematic reviewsCitation15,Citation17 reveal inconsistent results in the findings of Clostridiales and Clostridia. We observed decreased levels of Clostridium sp. (specie in the Clostridia class) in IBS. Clostridia class is observed to be at higher levels in IBSCitation19,Citation20 and IBS-DCitation21 compared to HCs and has been speculated to be a part of the pathogenesis in IBS due to the contribution of overactive bile acid secretionCitation21 and dysregulation of gut serotonin production.Citation22,Citation23 Clostridia class contains gut pathogens causing some of the most common clostridial infections in the gut,Citation24 such as Clostridium difficile infections which is a common cause of post-infectious IBS.Citation25 But some Clostridium species are commensals and might have potentials as probiotics.Citation26 Their cellular components and metabolites play a probiotic role primarily through strengthening the intestinal barrier and interacting with the immune system. Unique patterns of Clostridium species in gut can be shaped depending on our diets and physical state of bodyCitation26 and a balance of Clostridium spp. is needed to form a healthy gut. Therefore, both decreased and increased numbers might cause IBS symptoms.
Patients with IBS are a heterogenous group with different stool habits affecting the fecal and luminal microbiota.Citation27,Citation28 This makes the comparison of gut microbiota in IBS across studies challenging as the relative frequency of each subgroup is different in all studies. We should take this into consideration and investigate all subgroups.
In respect to differences in IBS subgroups and HCs, IBS-D had significantly higher levels of Proteobacteria, Pseudomonas spp. (phyla Proteobacteria), Dorea spp. (phyla Firmicutes) and Ruminococcus gnavus (phyla Firmicutes) and significantly lower levels of Bacteroides pectinophilus (phyla Bacteroidetes), Actinobacteria and Bifidobacteria (phyla Actinobacteria) compared to HCs. Ruminococcus gnavus was found to be increased in IBS-D in two studies. Two studies also found DI to be significantly higher in IBS-D, hence, one study found a non-significant increase compared to HCs. DI in IBS-C and IBS-M was found to be significantly increased in comparison to HCs.
The findings of Ruminococcus gnavus being increased in IBS-D are directly in line with previous findings. The bacterial marker is a member of the Clostridia class (Firmicutes phyla) and it has been suggested to play a pathogenic role in IBS-D.Citation29 Monocolonization of germ-free mice with Ruminococcus gnavus induced IBS-D like symptoms, including increased GI transit and colonic secretion by stimulating the production of peripheral serotonin.Citation29 Serotonin (5-HT) has been shown to modulate gut motility and hypersensitivity functions.Citation30 The study of Rajilic-Stojanovic et al 2011Citation20 indicated that phylotypes related to Ruminococcus gnavus are significantly increased in patients with IBS and is positively associated with IBS symptoms. Increased levels of Ruminococcus gnavus contributed to the increased dysbiosis in patients with IBS in the study of Casén et al 2015,Citation10 along with increased levels of Proteobacteria (Shigella/Escherichia) and Actinobacteria. Our findings also reveal Proteobacteria (phylum) being increased in IBS-D according to Iribarren et al 2022.Citation13 An increasing amount of data identifies Proteobacteria as a possible microbial signature of disease as it includes pathogenic bacteria with potential inflammation causing mechanisms.Citation31,Citation32 Increased levels in IBSCitation17,Citation33 and IBS-DCitation31,Citation34 are observed, and several studies has found the bacterial marker associated with IBSCitation17 and with individuals with high DI scores.Citation12 We also found Dorea spp. (Firmicutes phyla) at higher levels in IBS-D. Proteobacteria and Dorea spp. are observed to be increased in IBS-D together with excessive excreted bile acidCitation21 and higher levels of Streptococcus spp., Dorea spp. and Ruminococcus are reported to be associated with potentially harmful properties.Citation20 It has been proposed that Dorea spp. is associated with increased gas production and increased gut permeability, which is thought to contribute to IBS pathophysiology.Citation35 Altogether, our findings of increased levels of Ruminococcus gnavus and Proteobacteria in IBS-D seems to be supported by the literature.
Furthermore, we observed that Actinobacteria and Bifidobacteria (phylum Actinobacteria)Citation14 was decreased in IBS-D compared to HCs. A significant reduction in Actinobacteria has previously been observed in IBSCitation20 and IBS-D.Citation34 A systematic review showed that patients with IBS tend to have decreased levels of BifidobacteriaCitation15 and it is found a 1,5 decrease in Bifidobacteria in IBS compared to HCs and a negative association between Bifidobacterium spp. and abdominal pain in IBS.Citation20 Actinobacteria is one of the major phyla in the gut microbiota and is pivotal in the maintenance of gut homeostasis. Classes of this phylum are widely used as probiotics demonstrating beneficial effects in many pathological conditions,Citation36 especially Bifidobacterium is believed to exert positive health benefits on the hostCitation37 and has been commercially exploited as probiotics.Citation38 These characteristics of Actinobacteria and Bifidobacteria might explain the observation of low levels of Actinobacteria in IBS-D. However, increased level of Actinobacteria was shown to contribute to the dysbiosis of IBS in the study of Casén et al 2015,Citation10 making the interpretation inconclusive.
Recent evidence suggests that dysbiosis might play a role in the pathogenesis of IBSCitation1,Citation39 and a systematic review and meta-analysisCitation40 investigating gut microbial dysbiosis in patients with IBS concludes that IBS, including IBS-D and IBS-C, are characterized by gut microbiota dysbiosis. These findings are in line with the observations of dysbiosis in IBS, IBS-D, IBS-M, and IBS-C.
The above-mentioned studies have aimed to differentiate IBS and/or IBS subgroups from healthy controls based on fecal and/or gut microbiota composition but the results are inconclusive. When adding relevant variables (as Rome criteria, fecal transit time, gut microbiota metabolites and more), and using machine learning techniques the ability to distinguish IBS and/or IBS subgroups from healthy controls has been shown possible and might be a future method for diagnosing patients with IBS.Citation41–43
Limitations
The similarities and differences in fecal bacterial profile across studies using the GA-map Dysbiosis Test in patients with IBS compared to HCs might be explained by several reasons; patients and cohorts have different ethnicities, geographical sites, and different diets. Studies suggest that people from the same ethnicity share the same number of gut microbiota and indicate that the ethnicity selects for specific gut taxa, and diet is reported to have profound effects on the gut microbiota composition.Citation16,Citation44 Furthermore, although in the same group of patients, there are interindividual variations in the gut microbiota profile. Such variations are poorly understood, and appears to obscure the microbiota’s associations with patients health and impacts the microbiota-based diagnostics of disease.Citation16 The use of both Rome II, III and IV criteria for participants included in this systematic review might affect the outcome. Rome IV criteria differs from Rome III criteria in different ways. The most distinct difference is the change in the criteria of “abdominal pain or discomfort” in Rome III (and Rome II) to only include “abdominal pain” in Rome IV.Citation45 Although Rome II, III and IV was used in the five studies included, threeCitation11,Citation12,Citation14 studies and some of the participants of Casén et alCitation10 used Rome III criteria.
The GA-map Dysbiosis Test training and test cohort was included from Norway, Sweden, Denmark, and Spain, in an effort to achieve heterogeneity. Hence, they found a significantly different fecal bacterial profile in the Spanish cohort compared to the Scandinavian, and different bacterial markers contributing to the dysbiosis of the Spanish cohort.Citation10 The studies included in the present systematic review are conducted at different geographical sites and include cohorts with different ethnicities. Moreover, all participants in the studies were either told to obtain from a special diet or to have a normal Scandinavian diet without any major dietary change months prior to inclusion, or there were not reported any dietary inclusion or exclusion criteria. The diet at the time of inclusion naturally includes variation and were not reported to be controlled for in these studies. In respect to medications, as a minimum criterion, all studies did not allow the use of systemic antibiotics four weeks to six months prior to inclusion, and some studies excluded the use of immunosuppressive and/or glucocorticoids, or large doses of commercially available probiotics, and some, any other medications.
Additional limitations of this systematic review are the low number of studies (n = 5) included. Only one study investigated DI and one study investigated bacterial markers in IBS compared to HCs. Hence, three studies investigated the DI and fecal bacterial markers in IBS-D compared to HCs. In total there was a low volume of cases and controls. A higher number of females in respect to men was included in most of the studies. This was expected as IBS is reported to be more frequent in females.Citation46 Ahluwalia et al 2021Citation11 reported IBS patients to be significantly older than HCs.
Conclusion
This systematic review reveals inconsistent findings in respect to differences in bacterial markers between IBS and IBS subgroups with healthy controls in studies using the GA-map Dysbiosis Test. However, IBS-D was observed to have increased levels of Ruminococcus gnavus in two studies and IBS and IBS subgroups, especially IBS-D tend to have higher relative frequency of dysbiosis compared to HCs. The GA-map Dysbiosis Test is stated as a precise and easy in use method to identify dysbiosis in IBS and IBD. Comparing gut microbiota profiles in IBS patients across different ethnicities, geographical sites, sample sites, with different diets, medication use and with different methods will obscure the results. Hopefully, future research will take these aspects into account and investing fecal microbiota profile and dysbiotic differences in IBS and IBS subgroups compared to HCs. This will increase our understanding of gut microbiota’s associations with IBS pathophysiology and the clinical consequences of intestinal dysbiosis.
Disclosure
The authors report no conflicts of interest in this work.
Additional information
Funding
References
- Drossman DA. Functional gastrointestinal disorders: history, pathophysiology, clinical features and Rome IV. Gastroenterology. 2016;150(6):1262–1279.e2. doi:10.1053/j.gastro.2016.02.032
- Sperber AD, Bangdiwala SI, Drossman DA, et al. Worldwide prevalence and burden of functional gastrointestinal disorders, results of Rome foundation global study. Gastroenterology. 2021;160(1):99–114 e113. doi:10.1053/j.gastro.2020.04.014
- Mearin F, Lacy BE, Chang L, et al. Bowel Disorders. Gastroenterology. 2016:S0016–5085.
- Enck P, Aziz Q, Barbara G, et al. Irritable bowel syndrome. Nat Rev Dis Primers. 2016;2:16014. doi:10.1038/nrdp.2016.14
- Drossman DA, Morris CB, Schneck S, et al. International survey of patients with IBS: symptom features and their severity, health status, treatments, and risk taking to achieve clinical benefit. J Clin Gastroenterol. 2009;43(6):541–550. doi:10.1097/MCG.0b013e318189a7f9
- Corsetti M, Whorwell P. The global impact of IBS: time to think about IBS-specific models of care? Therap Adv Gastroenterol. 2017;10(9):727–736. doi:10.1177/1756283X17718677
- Chong PP, Chin VK, Looi CY, Wong WF, Madhavan P, Yong VC. The microbiome and irritable bowel syndrome - A review on the pathophysiology, current research and future therapy. Front Microbiol. 2019;10:1136. doi:10.3389/fmicb.2019.01136
- Piche T, Barbara G, Aubert P, et al. Impaired intestinal barrier integrity in the colon of patients with irritable bowel syndrome: involvement of soluble mediators. Gut. 2009;58(2):196–201. doi:10.1136/gut.2007.140806
- Barbara G, Cremon C, Carini G, et al. The immune system in irritable bowel syndrome. J Neurogastroenterol Motil. 2011;17(4):349–359. doi:10.5056/jnm.2011.17.4.349
- Casen C, Vebo HC, Sekelja M, et al. Deviations in human gut microbiota: a novel diagnostic test for determining dysbiosis in patients with IBS or IBD. Aliment Pharmacol Ther. 2015;42(1):71–83. doi:10.1111/apt.13236
- Ahluwalia B, Iribarren C, Magnusson MK, et al. A distinct faecal microbiota and metabolite profile linked to bowel habits in patients with irritable bowel syndrome. Cells. 2021;10(6):1459. doi:10.3390/cells10061459
- Vasapolli R, Schulz C, Schweden M, et al. Gut microbiota profiles and the role of anti-CdtB and anti-vinculin antibodies in patients with functional gastrointestinal disorders (FGID). Eur J Clin Invest. 2021;51(12):e13666. doi:10.1111/eci.13666
- Iribarren C, Nordlander S, Sundin J, et al. Fecal luminal factors from patients with irritable bowel syndrome induce distinct gene expression of colonoids. Neurogastroenterol Motil. 2022;34(10):e14390. doi:10.1111/nmo.14390
- Mazzawi T, Lied GA, Sangnes DA, et al. The kinetics of gut microbial community composition in patients with irritable bowel syndrome following fecal microbiota transplantation. PLoS One. 2018;13(11):e0194904. doi:10.1371/journal.pone.0194904
- Pittayanon R, Lau JT, Yuan Y, et al. Gut microbiota in patients with irritable bowel syndrome-A systematic review. Gastroenterology. 2019;157(1):97–108. doi:10.1053/j.gastro.2019.03.049
- Gaulke CA, Sharpton TJ. The influence of ethnicity and geography on human gut microbiome composition. Nature Med. 2018;24(10):1495–1496. doi:10.1038/s41591-018-0210-8
- Duan R, Zhu S, Wang B, Duan L. Alterations of gut microbiota in patients with irritable bowel syndrome based on 16S rRNA-targeted sequencing: a systematic review. Clin Transl Gastroenterol. 2019;10(2):e00012. doi:10.14309/ctg.0000000000000012
- Liu HN, Wu H, Chen YZ, Chen YJ, Shen XZ, Liu TT. Altered molecular signature of intestinal microbiota in irritable bowel syndrome patients compared with healthy controls: a systematic review and meta-analysis. Dig Liver Dis. 2017;49(4):331–337. doi:10.1016/j.dld.2017.01.142
- Gargari G, Taverniti V, Gardana C, et al. Fecal clostridiales distribution and short-chain fatty acids reflect bowel habits in irritable bowel syndrome. Environ Microbiol. 2018;20(9):3201–3213. doi:10.1111/1462-2920.14271
- Rajilic-Stojanovic M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology. 2011;141(5):1792–1801. doi:10.1053/j.gastro.2011.07.043
- Zhao L, Yang W, Chen Y, et al. A Clostridia-rich microbiota enhances bile acid excretion in diarrhea-predominant irritable bowel syndrome. J Clin Invest. 2020;130(1):438–450. doi:10.1172/JCI130976
- Mawe GM, Hoffman JM. Serotonin signalling in the gut--functions, dysfunctions and therapeutic targets. Nat Rev Gastroenterol Hepatol. 2013;10(8):473–486.
- Yano JM, Yu K, Donaldson GP, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161(2):264–276. doi:10.1016/j.cell.2015.02.047
- Borriello SP. Clostridial disease of the gut. Clinl Infect Dis. 1995;20(Supplement_2):S242–250. doi:10.1093/clinids/20.Supplement_2.S242
- Wadhwa A, Al Nahhas MF, Dierkhising RA, et al. High risk of post-infectious irritable bowel syndrome in patients with Clostridium difficile infection. Aliment Pharmacol Ther. 2016;44(6):576–582. doi:10.1111/apt.13737
- Guo P, Zhang K, Ma X, He P. Clostridium species as probiotics: potentials and challenges. J Anim Sci Biotechnol. 2020;11(1):24. doi:10.1186/s40104-019-0402-1
- Tottey W, Feria-Gervasio D, Gaci N, et al. Colonic transit time is a driven force of the gut microbiota composition and metabolism: in vitro evidence. J Neurogastroenterol Motil. 2017;23(1):124–134.
- Fukumoto S, Tatewaki M, Yamada T, et al. Short-chain fatty acids stimulate colonic transit via intraluminal 5-HT release in rats. Am J Physiol Regul Integr Comp Physiol. 2003;284(5):1269–1276. doi:10.1152/ajpregu.00442.2002
- Zhai L, Huang C, Ning Z, et al. Ruminococcus gnavus plays a pathogenic role in diarrhea-predominant irritable bowel syndrome by increasing serotonin biosynthesis. Cell Host Microbe. 2023;31(1):33–44 e35. doi:10.1016/j.chom.2022.11.006
- Kendig DM, Grider JR. Serotonin and colonic motility. Neurogastroenterol Motil. 2015;27(7):899–905. doi:10.1111/nmo.12617
- Bennet SM, Ohman L, Simren M. Gut microbiota as potential orchestrators of irritable bowel syndrome. Gut Liver. 2015;9(3):318–331.
- Jalanka-Tuovinen J, Salojarvi J, Salonen A, et al. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut. 2014;63(11):1737–1745. doi:10.1136/gutjnl-2013-305994
- Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24(6):521–530, e248. doi:10.1111/j.1365-2982.2012.01891.x
- Krogius-Kurikka L, Lyra A, Malinen E, et al. Microbial community analysis reveals high level phylogenetic alterations in the overall gastrointestinal microbiota of diarrhoea-predominant irritable bowel syndrome sufferers. BMC Gastroenterol. 2009;9(1):95. doi:10.1186/1471-230X-9-95
- Altomare A, Di Rosa C, Imperia E, Emerenziani S, Cicala M, Guarino MPL. Diarrhea predominant-irritable bowel syndrome (IBS-D): effects of different nutritional patterns on intestinal dysbiosis and symptoms. Nutrients. 2021;13(5):1506. doi:10.3390/nu13051506
- Binda C, Lopetuso LR, Rizzatti G, Gibiino G, Cennamo V, Gasbarrini A. Actinobacteria: a relevant minority for the maintenance of gut homeostasis. Dig Liver Dis. 2018;50(5):421–428. doi:10.1016/j.dld.2018.02.012
- O’Callaghan A, van Sinderen D. Bifidobacteria and their role as members of the human gut microbiota. Front Microbiol. 2016;7:925. doi:10.3389/fmicb.2016.00925
- Picard C, Fioramonti J, Francois A, Robinson T, Neant F, Matuchansky C. Review article: bifidobacteria as probiotic agents -- physiological effects and clinical benefits. Aliment Pharmacol Ther. 2005;22(6):495–512. doi:10.1111/j.1365-2036.2005.02615.x
- Marasco G, Cremon C, Barbaro MR, Stanghellini V, Barbara G. Gut microbiota signatures and modulation in irritable bowel syndrome. Microbiome Res Rep. 2022. doi:10.20517/mrr.2021.12
- Wang L, Alammar N, Singh R, et al. Gut microbial dysbiosis in the irritable bowel syndrome: a systematic review and meta-analysis of case-control studies. J Acad Nutr Diet. 2020;120(4):565–586. doi:10.1016/j.jand.2019.05.015
- Fukui H, Nishida A, Matsuda S, et al. Usefulness of machine learning-based gut microbiome analysis for identifying patients with irritable bowels syndrome. J Clin Med. 2020;9(8):2403. doi:10.3390/jcm9082403
- Tap J, Derrien M, Tornblom H, et al. Identification of an intestinal microbiota signature associated with severity of irritable bowel syndrome. Gastroenterology. 2017;152(1):111–123 e118. doi:10.1053/j.gastro.2016.09.049
- Shankar V, Reo NV, Paliy O. Simultaneous fecal microbial and metabolite profiling enables accurate classification of pediatric irritable bowel syndrome. Microbiome. 2015;3(1):73. doi:10.1186/s40168-015-0139-9
- Beam A, Clinger E, Hao L. Effect of diet and dietary components on the composition of the gut microbiota. Nutrients. 2021;13(8):2795. doi:10.3390/nu13082795
- Lacy BE, Patel NK. Rome criteria and a diagnostic approach to irritable bowel syndrome. J Clin Med. 2017;6(11):99. doi:10.3390/jcm6110099
- Kim YS, Kim N. Sex-gender differences in irritable bowel syndrome. J Neurogastroenterol Motil. 2018;24(4):544–558. doi:10.5056/jnm18082

