Abstract
Recent progress in the understanding of hepatitis C virus (HCV) biology and the availability of in vitro models to study its replication have facilitated the development of direct-acting antiviral agents (DAAs) that target specific steps in the viral replication cycle. Currently, there are three major classes of DAA in clinical development: NS3/4A protease inhibitors, NS5B polymerase inhibitors, and NS5A directed inhibitors. Several compounds thought to bind directly with NS5A are now in various clinical trial phases, including the most advanced, daclatasvir (BMS-790052), ledipasvir (GS-5885), and ABT-267. While many NS5A-targeted compounds demonstrate picomolar potency, the exact mechanism(s) of their action is still unclear. In the clinic, NS5A HCV inhibitors show promise as important components in DAA regimens and have multifunctionality. In addition to inhibiting viral replication, they may synergize with other DAAs, possibly by modulating different viral proteins, to help suppress the emergence of resistant viruses. Structure-based models have identified target interaction domains and spatial interactions that explain drug resistance for mutations at specific positions (eg, residues 93 and 31) within NS5A and potential binding partners. This review provides, insights into the unique complexity of NS5A as a central platform for multiple viral/host protein interactions, and possible mechanism(s) for the NS5A inhibitors currently undergoing clinical trials that target this nonstructural viral protein.
Introduction
Hepatitis C virus (HCV) is a global health burden, with approximately 170 million people (3% of the world’s population) estimated to be infected worldwide.Citation1 More than three million people contract HCV each year,Citation2 and while 15%–30% of all HCV infections clear spontaneously,Citation3 the remaining 70%–85% (an estimated 120–130 million) of infections will develop into chronic hepatitis, which can lead to steatosis, cirrhosis, and hepatocellular carcinoma.Citation4 Unfortunately, most are unaware of their infection – HCV-associated liver diseases may only manifest after decades in undiagnosed individuals – and can potentially transmit the virus to others, primarily through contaminated blood.Citation5 In addition, HCV reinfection after treatment has been reported, making vaccine development desirable.Citation6 Accordingly, the burden of HCV-associated disease is predicted to rise over the next 20 years.Citation7 In fact, in the US, HCV has now superseded human immunodeficiency virus type 1 (HIV-1) as the leading cause of mortality due to an infectious agent.Citation1
As a member of the Flaviviridae, the general replication cycle of HCV is similar to that of other viruses of this family and replicates entirely within the cytoplasm.Citation8 Because it does not establish latency, HCV is curable, although the mechanism by which it mediates persistence remains unclear. Among all recognized positive-strand ribonucleic acid (RNA) viruses, HCV is unique in its ability to establish a chronic infection.Citation9 The HCV genome consists of a 9.6 kb, positive-sense, single-stranded, enveloped RNA, which encodes three structural proteins (core, E1, and E2), the ion channel protein p7, and six nonstructural (NS) proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B).Citation10 Each of these proteins has a role in HCV entry, infection, replication, or maturation and is therefore a potential drug target.
HCV is highly heterogeneous, which can be an obstacle to the development of a universal treatment and a preventative vaccine. According to the World Health Organization, six major HCV genotypes and several subtypes have been identified throughout the world. Subtypes 1a/b account for approximately 70% of all infections in the US, Europe, China and Japan,Citation11 and the remainder are generally genotype 2, 3, and 4.Citation12 The HCV genotype strongly predicts the response to the currently approved HCV treatments.
Over the last decade, the standard of care comprised a dual-therapy regimen containing peginterferon alpha (PEG-IFN), given once per week as a subcutaneous injection, and ribavirin (RBV), a guanosine (ribonucleic) analog given orally twice daily. Individuals with HCV genotype 1 or 4 infection are less likely (40%–50%) to demonstrate a sustained viral response (SVR) with these treatments compared with individuals with genotype 2 or 3 disease (75%–85%).Citation13,Citation14 In 2011, the first NS3/4A HCV protease inhibitors (PIs), telaprevir and boceprevir, were approved. These direct-acting antiviral agents (DAAs) have now been licensed in several countries for use in combination with PEG-IFN and RBV, for the treatment of genotype 1 subjects. Unfortunately, many infected individuals, regardless of genotype, have been ineligible or unable to tolerate the standard of care regimen due to adverse effects and long treatment durations. Therefore, newer treatments with improved characteristics are needed to address the growing unmet medical needs.
HCV primarily infects liver parenchymal cells (hepatocytes). Because the liver is a highly specialized and complex organ, it is difficult to adequately model its biology in vitro. However, significant efforts have been directed at developing cell culture models to elucidate the viral replication in vitro.Citation15,Citation16 Specifically, the discovery of host cell receptor molecules that potentiate HCV infection has helped to overcome these obstacles, and the development of human hepatoma cell lines (eg, Huh-7 and Hep3B cells) has led to recent advances in the understanding of HCV structure and replication.Citation10,Citation15
There is now a broad pipeline of drugs in clinical development for treatment of HCV that relies on DAAs alone. DAAs block viral production by directly inhibiting one or more steps of the HCV replication cycle and are in various stages of clinical development.Citation13,Citation17–Citation24 DAAs currently include first-generation, second-wave, and second-generation NS3/4A PIs, nucleoside inhibitors and nonnucleoside inhibitors of the NS5B RNA polymerase, and NS5A complex inhibitors.Citation25–Citation28 Highly potent once-a-day, DAA-only combinations are now in Phase II and III of clinical development.Citation20 This review will focus on the unique complexity of NS5A as a drug target, with its putative protein interactions and possible mechanism(s) of action. We will summarize the most recent mechanistic research on drugs targeting NS5A and the ongoing clinical trials with these agents.
Structure and function of NS5A
NS5A is a 447 amino acid (aa), zinc-binding phosphoprotein comprised of three domains separated by two linker regions ().Citation29 Structurally, the amino-terminus of NS5A comprises the amphipathic α-helix, which is responsible for anchoring to the endoplasmic reticulum (ER) and ER-derived membranes, including lipid droplets (LDs).Citation30,Citation31
Figure 1 Domain organization schematic for the NS5A protein.
Notes: (A) D-I (aa 1–213), D-II (aa 250–342), and D-III (aa 356–447) are depicted with green, blue, and red boxes, respectively. Each domain is separated by an LCS (LCS-I and LCS-II). The N-terminal AH is depicted with a yellow box. D1a and 1bCitation33 are designated by dashed green lines. Domain numbering is from Tellinghuisen et al.Citation29 (B) Rows (1–5) show the sequence alignments and domain outlining of the HCV NS5A genotypes 1a, 1b, 2a, 3a, and 4a. Conserved and consensus residues are in red and black on the top line. The critical N-terminal, membrane binding, AH subdomain (aa 1–25) of D1 is outlined in gold. The critical conserved Cys residues that bind Zn++ in subdomain D1a are shaded blue. The proposed RNA-binding subdomain is D1b. Positions 28, 30, 31, and 93 (shaded red) are clinically observed mutations to NS5a-directed inhibitor treatment, which are also associated with high cell-based resistance. Rows (6–8) show the sequence of experimental 3D structures R7G.pdb (by NMR, aa 1–31), 3FQQ.pdb (by X-ray diffraction, aa 31–207), and 1ZH1.pdb (by X-ray diffraction, aa 25–198) based on genotypes 1a, 1b, and 1b, respectively.
Abbreviations: 3D, three-dimensional; aa, amino acid; AH, amphipathic helix; Cys, cysteine; HCV, hepatitis C virus; LCS, low complexity sequence; NMR, nuclear magnetic resonance; NS5A, nonstructural protein 5A; Pdb, protein data bank; RNA, ribonucleic acid; D, domain.
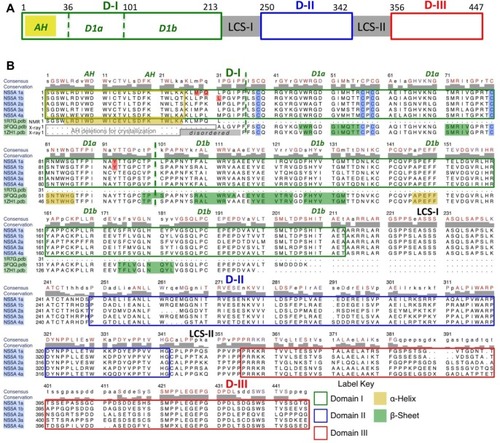
In one study, the structure encoded in the first 31 aas of HCV genotype 1a domain I (D-I) was determined, by nuclear magnetic resonance spectrometry (NMR), to include an extended amphipathic helix (AH), which oscillates in plane with the cytosolic surfaces of the model membrane bilayers and which is thought to be conserved for all genotypes ().Citation32 The remaining subdomains were subsequently crystallized using different truncated sequences of genotype 1b ().Citation33,Citation34 Both three-dimensional (3D) structures reveal a novel folded core between aa 36–100 designated as subdomain 1a (D1a), that coordinates a zinc atom via four cysteines, yet the two structures pack into two unique conformations of the homodimer. The remaining residues in D-I (aa 101–213), designated subdomain 1b (D1b), include a putative RNA-binding domain at the dimer interface of one of the structures.Citation33 The structural predictions were recently confirmed by in vitro biochemical assays, where the D-I residues involved in dimerization and RNA binding were identified.Citation35 Domains II (D-II) and III (D-III) of NS5A are intrinsically disordered and are therefore flexible. This may confer NS5A’s promiscuous ability to interact with numerous proteins.Citation36–Citation40 Specifically, D-II is involved with binding to cyclophilin A and, therefore, has the potential to antagonize the innate immune response to HCV.Citation41 D-III seems to be important for the assembly of infectious viral particles.Citation42,Citation43
NS5A exists in two forms (designated p56 and p58), based on electrophoretic mobility.Citation44 The p56 form is primarily unphosphorylated or is basally phosphorylated by several kinases in different regions,Citation45–Citation47 and data suggests that p58 hyperphosphorylation is performed by casein kinase I isoform alpha.Citation48 More recently, the essential host factor, lipid kinase phosphatidylinositol 4-kinase III alpha (PI4KIIIα) has been shown to bind NS5A and regulate its phosphorylation state.Citation49 Surprisingly, the upregulation of PI4KIIIα has been shown to result in lower levels of the hyperphosphorylated p58.Citation49 While the exact role of NS5A phosphorylation in regulating HCV RNA replication is not completely understood, it may contribute to a switch between viral RNA replication versus RNA translation and/or packaging.Citation50 Recent studies suggest that NS5A hyperphosphorylation appears to play different roles between in vitro and in vivo systems, and the degree and requirements for hyperphosphorylation may vary between different HCV genotypes and isolates.Citation51,Citation52
While NS5A is essential to HCV genome replication and is required for virion morphogenesis, its specific role in these processes has yet to be determined. What is certain is that HCV, like virtually all plus-strand RNA viruses, has the hallmark feature of forming a membrane-associated replication complex composed of virus proteins, replicating RNA and altered cellular membranes.Citation53,Citation54 All the HCV NS proteins, including NS5A, interact with host cell membranes, either directly as membrane-binding proteins, or in the case of NS3, via interaction with the membrane-anchoring protein NS4A.Citation16 The interaction of these viral proteins with the membrane forms tight structures that accumulate as a “membranous web” to house the viral replication complexes.Citation53
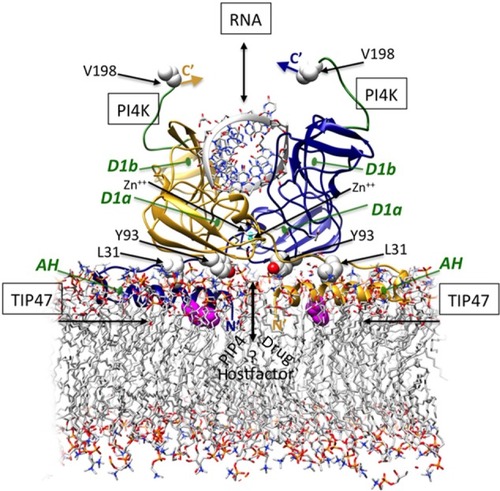
Even though NS5A has no known enzymatic activity, its interaction with the membranous web is critical for viral replication, and modification/deletion of specific residues of the membrane-binding AH domain impairs both ER localization and RNA replication.Citation30,Citation32,Citation55 NS5A interacts with a myriad of cellular and viral factors, including viral RNA, which likely enables it to perform multiple roles within the virus replication cycle. For example, NS5A is a novel structural class of RNA-binding protein with the ability to bind to HCV RNA.Citation56–Citation58 NS5A also colocalizes with other HCV-encoded and host proteins, which may facilitate virus particle productionCitation59 and mediate replication complex movement via an association with microtubules and actin filaments.Citation60,Citation61 A theoretical model of the membrane-bound NS5A D-I structure, with viral RNA as a platform for the assembly of replication complexes, is presented in .
Figure 2 Structure-based theoretical model of membrane-bound NS5A D1 homodimer binding RNA duplex.
Notes: The ribbon representations of the dimeric subunits are colored gold and blue. The N-terminal AHs align in plane with the membrane and pack against residues Y93 and L31 at the dimer interfaces. This AH conformation suggests potential binding site(s) for endogenous cofactors and drugs at the Y93/AH/dimer interfaces that may help explain the activity of NS5A direct inhibitors at different stages of replication and inhibition, and the effects on membranous web morphology. An atom of Zn++ (cyan) binds at the D1a site at the core of each monomer, stabilizing a novel fold. RNA is shown binding at the D1b interface of this dimeric form. The recently identified regions for PI4K-binding at the C-terminal are colored green (182–198), and critical TIP47-binding residues near the AH N-termini are colored magenta. (The templates used PDB ID IZH1, PDB ID IR7G, POPC phospholipid bilayer). Reproduced with permission of James H Nettles. Copyright © 2013.
Abbreviations: AH, amphipathic helix; D1, domain 1 subdomain 1; NS5A, nonstructural protein 5A; PI4K, phosphatidylinositol 4-kinase; PIP4, phosphatidylinositol phosphate 4; pdb, protein data bank; RNA, ribonucleic acid; TIP47.
In addition to forming viral protein complexes within the membranous web, evidence suggests that NS5A interacts with diverse host proteins. For example, NS5A has attracted considerable interest because of its potential role in modulating the response to interferon (IFN)-α therapy,Citation62 although this remains controversial. NS5A also alters intracellular events (eg, calcium levels), which leads to oxidative stress and the activation of the nuclear factor kappa-light-chain enhancer of activated B cells (NF-κβ) and signal transducer and activator of transcription (STAT-3) transcription factors in the liver.Citation63,Citation64 As previously referenced, NS5A was found to directly interact with PI4KIIIα and stimulate its kinase activity, which reduces p58 hyperphosphorylation.Citation49 Importantly, this leads to the formation of a phosphatidylinositol-4 phosphate (PI4P)-enriched membranous environment, which may be critical for the HCV replication cycle as the inhibition of PI4KIIIα abrogates HCV replication.Citation65 Also, PI4P has been shown to rearrange during HCV infection and to colocalize with NS5A.Citation66 Most recently, the cytoplasmic sorting factor tail-interacting protein of 47 kD (TIP47) was found to have a crucial role in the HCV replication cycle and to bind NS5A directly at the conserved AH residue tryptophan (W) 9.Citation67,Citation68 In short, while the specifics of the HCV replication cycle have yet to be fully understood, it is likely that NS5A functions as a key modulator of this process.
NS5A inhibitor mechanism(s) of action
A recent clinical study of nine HCV-infected persons who were administered 10 or 100 mg single doses of the NS5A inhibitor daclatasvir revealed a multiphasic decline in serum HCV.Citation69,Citation70 An initial, rapid two-log10 drop in HCV RNA levels over the first 6 hours and a half-life (t1/2) of 48 minutes was observed, suggesting a direct inhibition of virus assembly or the secretion in serum. This was followed by a much slower phase of decline (t1/2= 6–9 hours), which was indicative of an effect on viral RNA synthesis. These clinical findings suggest that NS5A inhibitors have mechanism(s) that affect both viral genome replication and the assembly/release of infectious HCV particles in humans, related to those previously seen in cell-based studies.
In vitro experiments indicate that NS5A is a “promiscuous” protein, interacting with multiple host and virus components, most likely involved in the membranous web formation.Citation53,Citation71 For instance, NS5A has been shown to interact with core protein to enhance the formation of HCV particlesCitation72–Citation74 and with NS3, NS4A, and NS5B to modulate its phosphorylation, in addition to its role in binding RNA.Citation33,Citation56,Citation57,Citation75–Citation77 However, it is not precisely clear which complex(es) are the target of NS5A-directed inhibitor drugs. Attempts at monitoring inhibition, in a cell-free system with only purified NS5A, have not been successful to date, suggesting that targeting the NS5A complex may require additional host and viral factors. On the other hand, the HCV replicon system has provided a rapid and efficient way to monitor the effects of NS5A inhibitors on HCV RNA production within the membranous web complex. It is noteworthy that the effect(s) of NS5A inhibitors on viral assembly and release cannot be studied directly using the replicon system, as replicon cells do not contain the envelope gene.
Recent mechanistic studies using immunofluorescence to track NS5A localization in replicon-containing cells following treatment with NS5A inhibitors have suggested that these drugs reduce viral RNA production in new, rather than preformed, replication complexes, which may correlate with the observed slow rate of viral load decay seen in clinical trials.Citation78,Citation79 These studies also demonstrated that NS5A inhibitors significantly shift the distribution of NS5A from the ER to LDs and that such redistribution is ablated in cells with drug-resistant mutations (eg, Y93H and L31V).Citation78 Since NS5A has previously been shown to colocalize with core proteins on LDs,Citation31,Citation80 drug binding at the LD stage may interfere with viral assembly and particle release at the cell membrane, thus accounting for the fast decay (as part of the multiphasic decline) seen in the clinic.
Additional mechanisms for the biphasic clinical response to NS5A-targeted inhibitors may include functional complexes with essential host factors, such as PI4KIIIα or TIP47. PI4KIIIα creates a PI4P-rich membranous environment supporting HCV replication as well as modulating NS5A phosphorylation.Citation49,Citation81 TIP47 is an LD-binding host protein that has previously been shown to regulate RNA replication via direct interaction with HCV NS5A and to have a key role in the viral assembly/release of the related dengue flavivirus.Citation82,Citation83
While we have described at least two distinct possible modes of action for the drug response observed in infected persons, these modes of action are not likely to be exclusive. If NS5A inhibitors only affect the formation of new viral replication complexes, this mode of action would be limited to a relatively narrow time period in the HCV replication cycle; however, if NS5A inhibitors additionally affect the virus assembly complex, preventing the maturation of viral particles and the spread of infection, these mechanisms would occur at two different stages of the replication cycle and could account for the observed multiphasic kinetics.Citation67,Citation68
Both mechanisms suggest that the ideal NS5A inhibitors should be administered at doses that maintain steady-state trough concentrations (Ctrough) that will maintain an SVR at week 12 (SVR12). Because it is assumed that only the unbound fraction of drug is free to diffuse into hepatocytes, many studies have reported maintenance of Ctrough > the in vitro determined 90% effective concentration (EC90) against the targeted strain of HCV, corrected for binding to plasma proteins (protein-corrected potency), as a biomarker of therapeutic efficacy (). Once-a-day antiviral regimens generally demonstrate a greater degree of compliance. Once-a-day formulations are more easily achieved using drugs that have a long plasma t1/2; eg, ledipasvir (t1/2=49.7 hours), ABT-267 (t1/2=28.1 hours), and daclatasvir (t1/2=12.8 hours). However, given the high potency of NS5A inhibitors, drugs with short t1/2 may also be considered as once-a-day regimens, provided a sufficiently high dose could be administered to maintain an effective steady-state trough without producing systemic toxicity. The pharmacokinetics of NS5A inhibitors in clinical development are summarized in .
Table 1 Pharmacokinetic parameters of NS5A inhibitors
Chemical space of NS5A-directed inhibitors
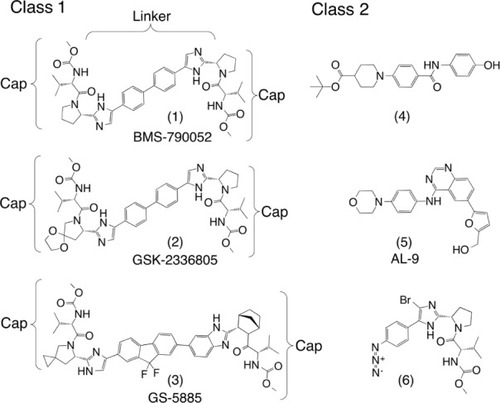
Daclatasvir was discovered using HCV replicon systems for high throughput screening (HTS), which identified a low micromolar hit that selected mutations in the NS5A coding region.Citation84 Further screening of structurally similar compounds identified a low nanomolar lead that selected the same mutations.Citation84 Investigation of the mechanism of the lead’s increased potency revealed a homodimeric metabolite to be the active component.Citation85 Further medicinal chemistry optimization of the symmetric, dimeric scaffolds resulted in the picomolar-active structure of BMS-790052, also known as daclatasvir.Citation85,Citation86 A recent review mapped the chemical space of over 50 compounds from the patent literature. They were found to center around daclatasvir’s chemical structure.Citation27 This large set of compounds can be grouped into two general chemotype classes (summarized in ). The class 1 compounds are dimers presenting a chemical motif of two peptidic caps joined by conjugated aromatic core linkers and include the known clinical compounds daclatasvir (BMS-790052), GSK-2336805, and ledipasvir (65-5885), with the structures shown in . The most common cap is the proline-valine-carbamate motif seen in these three, while the linkers can vary widely.Citation27 The class 2 compounds are distinct from class 1 and include monomers with very different chemical scaffolds. In general, the class 2 compounds are less active than Class 1 in replicon assays with nanomolar, rather than picomolar, median 50% effective concentration (EC50) values. “Compound 4,” with an EC50 of 160 nM, was the most potent, in vitro, of a series built on a piperazine-aryl scaffold from Merck and Co, Inc (Whitehouse Station, NJ, USA).Citation87 Mutations selected by this compound localize on NS5A, near the homodimer interface of the crystal structure, suggesting that effecting dimerization may be part of its molecular mechanism.Citation87 However, some other class 2 members clearly point to alternative mechanism(s) of action for NS5A-targeted compounds. For example, 4-anilino quinazoline (AL-9) from Arrow Therapeutics Limited (London, UK) was first thought to target NS5A, based on selected mutations that emerged in NS5A.Citation88 However, AL-9 was recently shown to directly inhibit purified PI4KIIIα and to reduce PI4P levels in the plasma membrane, which are known to regulate viral propagation.Citation89,Citation90
Figure 3 Representatives of the two general classes of NS5A-directed compounds found in the patent literature.
Notes: The dimeric class 1 compounds (1) (daclatasvir), (2), and (3) (ledipasvir) are the only three currently under clinical evaluation for which structures have been released. The class 2 compounds are diverse chemotypes and include monomers that do not fit class 1 (4–6).
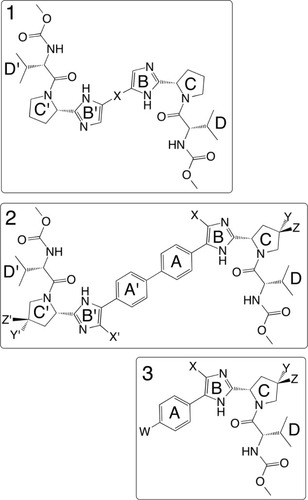
Systematic explorations of the structure–activity relationships (SARs) for compounds from both class 1 and class 2 were recently reported by our group, in collaboration with RFS Pharma, LLC (Tucker, GA, USA).Citation91–Citation93 Briefly, analog libraries of symmetric dimer compound 1, sharing a proline-valine-carbamate cap, were synthesized in three series (). Series 1 included replacement of the two “A” phenyl rings with shorter, nonaromatic linkers, but these substitutions resulted in >1,000-fold drop in potency. However, the straight aromatic linkers could be extended by up to five rings and still retain picomolar potency.Citation93 The addition of specific halogen substitution at ring “B” was able to rescue the potency of an inactive parent compound with bent linkers, which suggests a key role for conformational stability related to potency. For series 2, symmetric halogen substitutions (Cl, Br, I, or F) at the x and x’ positions produced compounds with similar EC50 to the parent compound. Surprisingly, an aryl substitution at the imidazole region B maintained activity at 200 pM, and the oxaryl group improved potency to 26 pM, suggesting an open binding pocket in that region. In addition, an azido group substitution of the proline-like ring “C” at position y or z did not alter the activity (~20 pM), while larger 1,4 triazoles were only tolerated at position z, suggesting stereochemical restraints of the binding pocket.Citation92 Series 3 explored class 2 monomer activity. While the nonsubstituted scaffold was found to be completely inactive, a dual substitution with an azido group at position w and a bromo group at position x produced a highly active monomer, compound 6, with picomolar inhibition of genotype 1b replicons ().Citation91 Resistance selection with both class 1 and 2 compounds in this series identified the emergence of NS5A mutations at positions 93 and 31, and modeling has also supported the potential of shared binding site(s) for highly active members of both classes (Schinazi, unpublished data, 2013).
Figure 4 Three chemical series exploring systematic substitutions of a common compound 1 monomeric “cap” and relating specific features associated with activity across both class 1 (dimer) and class 2 (monomer) NS5a directed inhibitors.
Researchers at Pharmasset Inc (now Gilead Sciences, Foster City, CA, USA) used computational modeling to investigate the key pharmacophore responsible for the high activity of the class 1 dimers and theorized the importance of an intramolecular hydrogen bond between regions B and C to stabilize a conformation resembling a peptide “gamma-turn.”Citation94 Their development of a low picomolar active fluoroolefin-based mimetic supports that hypothesis and may provide a structural clue about the biological site of action. Although daclatasvir is both symmetric and dimeric by design, newer asymmetric and monomeric analogs have also been found to have high activity, suggesting that symmetry and high molecular weight dimers may not be a critical requirement for productive interactions with NS5A, in the next-generation compounds.
Resistance and biological space of NS5A-directed inhibitors
Overcoming acquired resistance is a major challenge for all antiviral targets, but for NS5A, acquired resistance defines the target. Daclatasvir, for example, was discovered using HCV replicon systems, which identified the rapid emergence of key resistance-bearing mutations at residues 31 and 93 in the NS5A coding region for genotypes 1a and 1b.Citation84 Site-directed mutagenesis at those positions resulted in significant loss of drug activity and correlated with selection of the same mutations in clinical trials.Citation86,Citation95,Citation96 The genotype 1b mutations Y93H and L31V are striking examples of aa changes that individually increase resistance 24- and 28-fold, respectively, but which increase resistance 14,789-fold when combinedCitation97 (), suggesting a specific biological site for drug binding. The locations of these drug-resistant mutations within the AH-D1a linker and near the D1a/D1b interface are highlighted for genotype 1b in .
Table 2 Resistance profile and fitness of daclatasvir, in genotype 1a and 1b replicon systemsCitation97
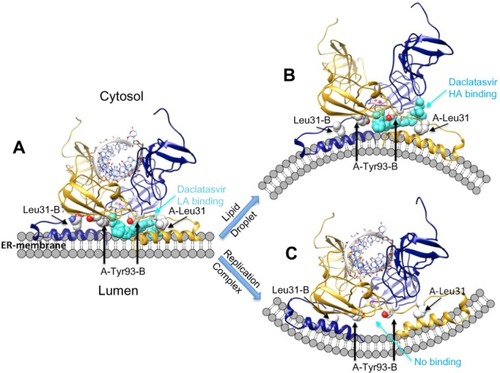
The structure of aa 1–31 at the N-terminal of NS5A was elegantly solved by NMR in lipid bilayer models. It was found to form a flexible amphipathic α-helix between residues 5–25 that inserted along the membrane surface with hydrophobic residues buried, polar residues exposed, and amphiphilic tryptophan residues aligned in plane (PDB ID 1R7G).Citation32 The two 3D X-ray crystal structure solutions for subdomains D1a and D1b reveal very different dimer alignments ().Citation33,Citation34 Because the deletion of AH was required for crystallization in each case, computational modeling has been employed to visualize the potential drug binding region at the AH–D1a interface. Cordek et al reported the use of a full-length NS5A AH-D1 homology model (combining the α-helix from NMR with the dimer from PDB ID 3FQQ) to dock both class 1 and class 2 NS5A inhibitor compounds developed at Glaxo-SmithKline, PLC (Brentford, UK).Citation27 While residues 28, 30, and 31 were described to be in close proximity to Tyr 93 at the dimer interface of the model, the direct binding mode was reported as “elusive.”Citation27 More recently, a group from Bristol-Myers Squibb (New York, NY, USA) has proposed a symmetric binding mode at the dimer interface of their PDB ID 1ZH1-based structure; however, it does not easily explain the high activity of some class 2 compounds.Citation98 A new structural review has also reported the potential for connecting AH from NMR to either of the X-ray crystal structures, but with ambiguity of the drug-binding detail.Citation28 Our group has studied both dimer forms as platforms for building full-length NS5A AH–D-I homology models and tested multiple possible AH–D1a linker conformations related to drug binding. We found the PDB ID 1ZH1 dimer template to provide better AH alignments, based on chemical genetics-derived distance constraints (J Nettles, unpublished data, 2013). Our analysis for docking NS5A inhibitors to both genotype 1a and 1b models predicts the asymmetric drug binding of one cap between the pair of Tyr 93 residues at the N-terminal AH/dimer/membrane interface, with a secondary cap interaction between 93 and 31 of the two monomers (). This binding mode can accommodate both class 1 and class 2 binding (J Nettles, unpublished data, 2013). These results further suggest a potential for conformational changes in the AH linker region that allows higher affinity asymmetric binding of the class 1 dimers to a membrane morphology associated with LDs (). The “open” 93/31 site allows the second cap to penetrate closer to the D1a core packing near residue 30, consistent with the resistance data of and the LD sequestering mechanism suggested by Targett-Adams et al.Citation78 Also consistent with the Targett-Adams results, the AH conformation associated with the replication complex does not support binding at either the 93/93 or 93/31 interfaces, due to a modified shape at the membrane interface (). Of note, these models of binding () coincide with the BMS results reported by O’Boyle et al that a photoactive analog of daclatasvir was found to covalently bind a NS5A peptide spanning aa 21–30.Citation98
Figure 5 Theoretical models of daclatasvir (turquoise) binding to NS5A and potential AH mediated effect on membrane morphology.
Notes: The models suggest the drug (turquoise) binds simultaneously to two asymmetric sites at the NS5A A/B dimer/membrane interface (blue/gold). A core site between Y93 of each monomer binds one cap of the drug, while the second cap fits between residues 93 and 31 of the aligned subunits. (A) The aromatic linker provides favorable interactions and positions the caps simultaneously within the two LA sites formed by AH aligned in the membrane plane. (B) Conformational change of AH exposes an HA “open” drug site between 93 and 31 of the different subunits. Binding to this state may lock NS5A into a conformation conducive to lipid droplet formation and release to cytosol, and is thought to impair assembly of other viral oligomers. (C) NS5A binding with RNA, NS5B, and other proteins induces replication complex formation in a membranous web and significantly lowers the affinity for drug binding. Reproduced with permission of James H Nettles. Copyright © 2013.
Abbreviations: AH, amphipathic helix; HA, higher affinity; LA, lower affinity; Leu, leucine; NS5A, nonstructural protein 5A; RNA, ribonucleic acid; Tyr, tyrosine.
NS5A inhibitors in clinical trials
According to http://www.clinicaltrials.gov, at least 13 potent NS5A DAAs have been evaluated in clinical trials (see ). Among them, three drugs are currently in Phase III clinical trials, including daclatasvir, ledipasvir, and ABT-267; seven drugs are in Phase II, and three drugs are in Phase I. To date, only three structures have been released and are summarized in .
Table 3 NS5A agents in clinical development for chronic HCV infection
Drugs currently in Phase III clinical trials
Daclatasvir/BMS-790052
Daclatasvir/BMS-790052 (Bristol-Myers Squibb) is the first-in-class NS5A inhibitor of the HCV replication complex, which in humans, has demonstrated a rapid and robust HCV RNA decline (3.6 log10), with no signs of adverse effects.Citation99 This compound blocks NS5A hyperphosphorylation and has the potential of shifting the subcellular localization of the viral protein.Citation78,Citation97 Daclatasvir has been shown to be highly effective against genotypes 1 and 2, and when in combination with IFN plus RBV, daclatasvir demonstrated a 100% SVR at 12 weeks posttreatment in persons infected with genotype 4. Impressively, in a Phase II study using IFN-free regimens with or without RBV, daclatasvir plus sofosbuvir (GS-7977) demonstrated a 100% SVR at 4 weeks (SVR4) and a 100% SVR12 in genotype 1–infected subjects who were either treatment naïve or who had failed prior treatment with telaprevir or boceprevir plus PEG-IFN/RBV, and 91% for genotypes 2– and 3–infected persons.Citation100 Furthermore, when combined with NS3PI, this was the first compound to demonstrate the proof of concept that an IFN-free regimen could eradicate HCV.Citation101 Based on the Phase II study results, which look promising, Phase III studies of the use of daclatasvir in combination with asunaprevir in genotype 1b treatment-naïve individuals are currently ongoing, and studies of the combination with asunoprevir and BMS-791325 are planned to start shortly.Citation102
Ledipasvir/GS-5885
Ledipasvir/GS-5885 (Gilead) demonstrated a high potency for HCV genotypes 1a (34 pM), 1b (4 pM), 4a (110 pM), and 6a (110 pM) but lower activity against genotypes 2a (21 nM) and 3a (41 nM), with an excellent overall selectivity index (>800,000 in replicon cells).Citation103 A Phase I clinical studyCitation104 of two doses showed a 2.3 and 3.3 log10 HCV genotype 1 load reduction after 3 days of monotherapy (1 and 10 mg QD [daily], respectively) but with quick emergence of resistance (residues 30 and 31 in genotype 1a and residue 93 in genotype 1b). During a first Phase II trial (ELECTRON), the combination of ledipasvir, sofosbuvir, and RBV led to 100% SVR for genotype 1 after 12 weeks, in treatment-naïve individuals and prior nonresponders. Overall, the regimen was well tolerated: anemia (20%), depression (8%), and headache (4%).Citation105 A Phase II trial (LONESTAR) involving the combination of ledipasvir (QD) and sofosbuvir (QD) without RBV, for 12 weeks led to SVR4 in 19/19 treatment-naïve and in 18/19 treatment-experienced genotype 1 subjects.Citation106 In another study, the combination of ledipasvir (30 mg QD) with PEG-IFN (180 mg/week) plus RBV (1,000–1,200 μg/day) with or without GS-9451 (a NS3 PI, 200 mg QD) led to a high SVR4 in treatment-naïveCitation107 and treatment-experiencedCitation108 subjects infected with HCV genotype 1. The same combination led also to high SVR12 rates in treatment-naïve, IL28B CC patients, which were comparable with the results of 24 weeks treatment with PEG-IFN plus RBV.Citation109 The four-drug therapy (ledipasvir plus GS-9451 plus PEG-IFN plus RBV) was generally safe and well tolerated; however, treatment was discontinued due to a severe pancytopenia case.Citation108,Citation109 In October 2012, Gilead initiated a combination study called ION-1 in a treatment-naïve genotype 1 HCV-infected population, with fixed doses (QD) for 12 or 24 weeks of ledipasvir and sofosbuvir, with and without RBV, which led to SVR4 >60%, with no significant safety issues.Citation110 A second follow-up study (ION-2) was initiated (January 2013) involving treatment of nonresponders infected with the HCV genotype 1, with fixed doses (QD) of ledipasvir and sofosbuvir, with RBV (12 weeks) or with/without RBV (24 weeks).Citation110 A third Phase III trial was initiated in May, 2013 (ION-3) comparing 8 weeks duration with 12 weeks duration, with fixed doses (QD) of ledipasvir and sofosbuvir with and without RBV, in 600 noncirrhotic, treatment-naïve HCV genotype 1–infected subjects.Citation106
ABT-267
A Phase III clinical trial involving the combination of three DAAs plus a protease booster and RBV is currently underway. In a Phase IIb clinical trial with a quintuple twice-a-day drug combination of ABT-267 (AbbVie, North Chicago, IL, USA) plus the PI ABT-450 plus ritonavir plus the nonnucleoside inhibitor ABT-333 plus RBV, a SVR12 of 98% and 93% was achieved in treatment-naïve persons and null responders infected with HCV genotype 1, respectively ().Citation111
Drugs currently in Phase II clinical trials
ACH-3102
ACH-3102 (Achillion Pharmaceuticals) is a potent second-generation NS5A inhibitor with a demonstrated high pharmacologic barrier to resistance and a low potential for the emergence of resistant variants in genotype 1 replicon systems. In a Phase Ia clinical trial, ACH-3102 demonstrated a rapid and robust HCV RNA decline (3.6–4.6 log10 range) in genotype 1–infected subjects.Citation107 A Phase II clinical trial of ACH-3102 with RBV has been initiated ().Citation100
Samatasvir (IDX719)
Samatasvir (Idenix Pharmaceuticals, Inc, Cambridge, MA, USA) has been shown to be highly potent against genotypes 1–4 in vitro; however, a low-resistance barrier to the Y93H variant has been demonstrated with this NS5A inhibitor. Nevertheless, a Phase II clinical trial is planned with samatasvir plus two other DAAs (TMC647055, a nonnucleoside polymerase inhibitor; and simeprevir, a PI) from Janssen (Janssen Pharmaceuticals, Inc, Titusville, NJ, USA).Citation112
MK-8742
MK-8742 (Merck) is a highly potent compound against genotype 1. It appears to function as a disruptor of the replication complex. In combination with MK-5172 (a PI), this candidate drug exhibited a high barrier to the development of escape mutants.Citation113 Phase II clinical trials are underway.
AZD-7295/A-831
AZD-7295 (AstraZeneca, London, UK; Arrow Pharmaceutical, Inc) initially demonstrated promising potency against genotypes 1b and 1a replicons (EC50=7 nM and 1.24 μM, respectively).Citation114 In 2008, a Phase II trial of AZD7295 was initiated. The status of this compound remains unknown.
GSK2336805
GSK2336805 (AstraZeneca/Arrow) is a highly potent NS5A inhibitor displaying a low EC50 for genotypes 1a (44 pM), 1b (8 pM), 4a (1.8 pM), 5a (2.5 pM), and 6ab (9.1 pM) but less potency for genotypes 2a, 3a, 6c-g, and 6h-n isolates. A Phase I clinical trial showed that mild headache (8%) was the only observed side effect after 7 and 14 days treatment with single (10, 30, or 60 mg) and multiple (10, 30, or 75 mg) oral doses in healthy adults. GSK2336805 resulted in a 0.93 to 3.9 log10 load reduction after monotherapy (1 mg or 60 mg QD, respectively) in treatment-naïve HCV genotype 1 subjects.Citation115 Results of the Phase II, 12-week study involving a combination of GSK2336805 and VX-135 (uridine nucleotide prodrug, a NS5B polymerase inhibitor), with and without RBV, in treatment-naïve genotype 1 subjects have not been disclosed yet.Citation116
PPI-668
The Presidio (Presidio Pharmaceuticals, Inc, San Francisco, CA, USA) second-generation NS5A inhibitor, PPI-668, showed potent pangenotypic activity, with an EC50 range between 0.02 and 1.3 nM in replicon assays for HCV genotypes 1–7. During a Phase I, 3-day monotherapy study, PPI-668 resulted in a 3.5–3.7 and 3.01 log10 load reduction in genotypes 1 and 2–3 treatment-naïve persons, respectively. The treatment was well tolerated in both healthy volunteers (80–320 mg QD) and HCV-positive subjects (40–240 mg QD).Citation117 A Phase IIa combination study with BI201335 (faldaprevir), a PI, and BI207127, a nonnucleoside HCV polymerase inhibitor, with or without ribavirin is planned.
GS-5816
GS-5816 (Gilead) is a potent second-generation NS5A inhibitor displaying low EC50 values (7–59 pM) against genotypes 1–6. GS-5816 has also shown a high-resistance barrier and retains potency against common NS5A polymorphisms and resistance mutations (EC50=130 pM against the L31M mutation and 121 pM against the Y93C mutation). In addition, GS-5816 was shown to remain active against mutations from other classes of HCV inhibitors and to show additive to moderately synergistic activities when combined with other anti-HCV agents (including sofosbuvir).Citation118,Citation119 A first-in-human, 7-day, Phase I study with healthy volunteers showed that GS-5816 was well tolerated at doses varying from 5 to 450 mg. Gilead is now moving on to a 12-week, all oral Phase II study to evaluate GS-5816 (25 and 100 mg) in combination with sofosbuvir with and without RBV in treatment naïve persons with chronic genotype 1–6 HCV infection.Citation120
Drugs currently in Phase I clinical trials
ACH-2928
ACH2928 (Achillion Pharmaceuticals, New Haven, CT, USA) is a highly potent NS5A inhibitor. In a 3-day, monotherapy Phase I clinical trial, this compound demonstrated an impressive and rapid reduction of HCV RNA levels (3.7 log10) in persons infected with genotype 1.Citation121
EDP-239
EDP-239 (Enanta Pharmaceuticals, Inc, Watertown, MA, USA; Novartis Pharmaceuticals Corp, Basel, Switzerland) is a potent NS5A-targeted molecule, which in vitro, has demonstrated picomolar activity against genotypes 1b (EC50=7 pM) and 1a (EC50=31 pM). The combination of EDP-239 with the cyclophilin inhibitor alisporivir resulted in a strong synergistic anti-HCV effect against genotypes 1–3.Citation27,Citation122 Novartis has recently initiated Phase I clinical trials of EDP-239.
PPI-461
PPI-461 (Presidio Pharmaceuticals, Inc), Presidio’s first-generation NS5A inhibitor has shown potent in vitro activity for the HCV genotypes 1–6 and up to a 3.6 log10 load reduction after 3 days of monotherapy (100 and 200 mg oral doses) in HCV genotype 1–infected subjects.Citation123 However, a rapid emergence of resistance was observed (at residues 28, 30, 31, and 93). Presidio’s second-generation NS5A inhibitor, PPI-668, also has pangenotypic activity (described above).
Conclusion
Despite the current gaps in our understanding of NS5A, this NS protein has been implicated as an important modulator of critical viral functions, including viral RNA binding and replication, virus assembly, and regulation of the antiviral IFN response. Together, these roles present a unique platform for NS5A inhibitors, to be combined with more traditional target-based drug discovery. NS5A-directed compounds are considered to be the most potent anti-HCV molecules ever discovered. However, their Achilles heel is that they select drug-resistant mutants fast and to be effective, must be used with other potent DAA drugs, to prevent clinical resistance. Structure-based models have demonstrated the potential for identifying target domains and spatial interactions that rationalize the selection of resistance mutations at specific positions within NS5A and potential binding partners (eg, residues 93 and 31). Continued SAR studies will further elucidate the mechanism(s) of action of these small molecules and facilitate the discovery of new drugs with higher barrier to resistance.
NS5A-targeted therapies demonstrate a remarkable capacity to inhibit HCV RNA replication, with minimal in vivo toxicity. At least two molecules (daclatasvir and ledipasvir) are being clinically evaluated in IFN-free combination therapies. NS5A inhibitors appear to be emerging as important components of HCV DAA regimens with dual functionality. In addition to having picomolar potency, they can synergize with other DAA, targeting different viral proteins, to prevent and suppress the emergence of resistant viruses. If NS5A inhibitors can exercise their remarkable potency to rapidly drive down virus replication within the protection offered by other DAAs, then resistance to any one DAA component may be suppressed by other components in the combination regimens. Thanks to these advances, the future is looking promising for the millions of HCV-infected individuals. An IFN-free, oral, once-daily, pangenotypic drug combination of a nucleoside analog, such as sofosbuvir, with a next-generation pangenotypic NS5A drug, such as GS-5816, may play a pivotal role and could potentially cure millions of chronically infected HCV individuals and result in global HCV eradication in the near future.
Acknowledgments
This work was supported in part by National Institutes of Health grant 5P30-AI-50409 (Centers for AIDS Research) and by the Department of Veterans Affairs. The authors thank Judy Matthew for her careful editing of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
- LavanchyDThe global burden of hepatitis CLiver Int200929Suppl 1S74S81
- AlterMJEpidemiology of hepatitis C virus infectionWorld J Gastroenterol200713172436244117552026
- LehmannMMeyerMFMonazahianMTillmannHLMannsMPWedemeyerHHigh rate of spontaneous clearance of acute hepatitis C virus genotype 3 infectionJ Med Virol200473338739115170633
- TongMJel-FarraNSReikesARCoRLClinical outcomes after transfusion-associated hepatitis CN Engl J Med199533222146314667739682
- AlbertiAChemelloLBenvegnùLNatural history of hepatitis CJ Hepatol199931Suppl 1S17S24
- LambersFAPrinsMThomasXMOSAIC (MSM Observational Study of Acute Infection with hepatitis C) study groupAlarming incidence of hepatitis C virus re-infection after treatment of sexually acquired acute hepatitis C virus infection in HIV-infected MSMAIDS20112517F21F2721857492
- Deuffic-BurbanSPoynardTSulkowskiMSWongJBEstimating the future health burden of chronic hepatitis C and human immunodeficiency virus infections in the United StatesJ Viral Hepat200714210711517244250
- LohmannVHepatitis C virus RNA replicationCurr Top Microbiol Immunol201336916719823463201
- MoorePSChangYWhy do viruses cause cancer? Highlights of the first century of human tumour virologyNat Rev Cancer2010101287888921102637
- MoradpourDPeninFHepatitis C virus proteins: from structure to functionCurr Top Microbiol Immunol201336911314223463199
- KimAISaabSTreatment of hepatitis CAm J Med2005118880881516084169
- Wartelle-BladouCLe FolgocGBourlièreMLecomteLHepatitis C therapy in non-genotype 1 patients: the near futureJ Viral Hepat201219852553622762136
- AlexopoulouAPapatheodoridisGVCurrent progress in the treatment of chronic hepatitis CWorld J Gastroenterol201218426060606923155334
- ZeinNNRakelaJKrawittELReddyKRTominagaTPersingDHHepatitis C virus genotypes in the United States: epidemiology, pathogenicity, and response to interferon therapy. Collaborative Study GroupAnn Intern Med199612586346398849147
- WilsonGKStamatakiZIn vitro systems for the study of hepatitis C virus infectionInt J Hepatol2012201229259123056952
- MoradpourDPeninFRiceCMReplication of hepatitis C virusNat Rev Microbiol20075645346317487147
- BarreiroPVispoEPovedaEFernández-MonteroJVSorianoVHepatitis C therapy: highlights from the 2012 annual meeting of the European Association for the Study of the LiverClin Infect Dis201356456056623090932
- AssisDNLimJKNew pharmacotherapy for hepatitis CClin Pharmacol Ther201292329430522850602
- ShahNPierceTKowdleyKVReview of direct-acting antiviral agents for the treatment of chronic hepatitis CExpert Opin Investig Drugs201322911071121
- KiserJJFlexnerCDirect-acting antiviral agents for hepatitis C virus infectionAnnu Rev Pharmacol Toxicol20135342744923140245
- JesudianABGambarin-GelwanMJacobsonIMAdvances in the treatment of hepatitis C virus infectionGastroenterol Hepatol (NY)20128291101
- YangPLGaoMLinKLiuQVillarealVAAnti-HCV drugs in the pipelineCurr Opin Virol20111660761622440918
- De ClercqEThe race for interferon-free HCV therapies: a snapshot by the spring of 2012Rev Med Virol201222639241122936636
- FarnikHZeuzemSNew antiviral therapies in the management of HCV infectionAntivir Ther201217577178322626842
- HuntDPockrosPWhat are the promising new therapies in the field of chronic hepatitis C after the first-generation direct-acting antivirals?Curr Gastroenterol Rep201315130323250703
- BeldaOTargett-AdamsPSmall molecule inhibitors of the hepatitis C virus-encoded NS5A proteinVirus Res20121701–211423009750
- CordekDGBechtelJTMaynardATKazmierskiWMCameronCETargeting the NS5A protein of HCV: an emerging optionDrugs Future201136969171123378700
- BartenschlagerRLohmannVPeninFThe molecular and structural basis of advanced antiviral therapy for hepatitis C virus infectionNat Rev Microbiol201311748249623748342
- TellinghuisenTLMarcotrigianoJGorbalenyaAERiceCMThe NS5A protein of hepatitis C virus is a zinc metalloproteinJ Biol Chem200427947485764858715339921
- BrassVBieckEMontserretRAn amino-terminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5AJ Biol Chem2002277108130813911744739
- ShiSTPolyakSJTuHTaylorDRGretchDRLaiMMHepatitis C virus NS5A colocalizes with the core protein on lipid droplets and interacts with apolipoproteinsVirology2002292219821011878923
- PeninFBrassVAppelNStructure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5AJ Biol Chem200427939408354084315247283
- TellinghuisenTLMarcotrigianoJRiceCMStructure of the zinc-binding domain of an essential component of the hepatitis C virus replicaseNature2005435704037437915902263
- LoveRABrodskyOHickeyMJWellsPACroninCNCrystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virusJ Virol20098394395440319244328
- LimPJChatterjiUCordekDCorrelation between NS5A dimerization and hepatitis C virus replicationJ Biol Chem201228736308613087322801423
- GuptaGQinHSongJIntrinsically unstructured domain 3 of hepatitis C Virus NS5A forms a “fuzzy complex” with VAPB-MSP domain which carries ALS-causing mutationsPLoS One201276e3926122720086
- HanoulleXBadilloAVerdegemDPeninFLippensGThe domain 2 of the HCV NS5A protein is intrinsically unstructuredProtein Pept Lett20101781012101820450484
- HughesMGriffinSHarrisMDomain III of NS5A contributes to both RNA replication and assembly of hepatitis C virus particlesJ Gen Virol200990Pt 61329133419264615
- LiangYYeHKangCBYoonHSDomain 2 of nonstructural protein 5A (NS5A) of hepatitis C virus is natively unfoldedBiochemistry20074641115501155817880107
- Ross-ThrieplandDAmakoYHarrisMThe C terminus of NS5A domain II is a key determinant of hepatitis C virus genome replication, but is not required for virion assembly and releaseJ Gen Virol201394Pt 51009101823324467
- AnsariIUStrikerRSubtype specific differences in NS5A domain II reveals involvement of proline at position 310 in cyclosporine susceptibility of hepatitis C virusViruses20124123303331523342381
- AppelNZayasMMillerSEssential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assemblyPLoS Pathog200843e100003518369481
- KimSWelschCYiMLemonSMRegulation of the production of infectious genotype 1a hepatitis C virus by NS5A domain IIIJ Virol201185136645665621525356
- KanekoTTanjiYSatohSProduction of two phosphoproteins from the NS5A region of the hepatitis C viral genomeBiochem Biophys Res Commun199420513203267999043
- KimJLeeDChoeJHepatitis C virus NS5A protein is phosphorylated by casein kinase IIBiochem Biophys Res Commun1999257377778110208859
- ChenYCSuWCHuangJYPolo-like kinase 1 is involved in hepatitis C virus replication by hyperphosphorylating NS5AJ Virol201084167983799320534861
- KatzeMGKwieciszewskiBGoodlettDRSer(2194) is a highly conserved major phosphorylation site of the hepatitis C virus nonstructural protein NS5AVirology2000278250151311118372
- QuintavalleMSambuciniSDi PietroCDe FrancescoRNeddermannPThe alpha isoform of protein kinase CKI is responsible for hepatitis C virus NS5A hyperphosphorylationJ Virol20068022113051131216943283
- ReissSHarakCRomero-BreyIThe lipid kinase phosphatidylinositol-4 kinase III alpha regulates the phosphorylation status of hepatitis C virus NS5APLoS Pathog201395e100335923675303
- HuangYStaschkeKDe FrancescoRTanSLPhosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication?Virology200736411917400273
- LemayKLTreadawayJAnguloITellinghuisenTLA hepatitis C virus NS5A phosphorylation site that regulates RNA replicationJ Virol20138721255126023115292
- AppelNPietschmannTBartenschlagerRMutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domainJ Virol20057953187319415709040
- EggerDWölkBGosertRExpression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complexJ Virol200276125974598412021330
- GosertREggerDLohmannVIdentification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic repliconsJ Virol20037795487549212692249
- ElazarMCheongKHLiuPGreenbergHBRiceCMGlennJSAmphipathic helix-dependent localization of NS5A mediates hepatitis C virus RNA replicationJ Virol200377106055606112719597
- HuangLHwangJSharmaSDHepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding proteinJ Biolog Chem2005280433641736428
- HwangJHuangLCordekDGHepatitis C virus nonstructural protein 5A: biochemical characterization of a novel structural class of RNA-binding proteinsJ Virol20108424124801249120926572
- Targett-AdamsPBoulantSMcLauchlanJVisualization of double-stranded RNA in cells supporting hepatitis C virus RNA replicationJ Virol20088252182219518094154
- JiraskoVMontserretRLeeJYStructural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assemblyPLoS Pathog2010612e100123321187906
- LaiCKJengKSMachidaKLaiMMAssociation of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5AJ Virol200882178838884818562541
- LaiCKJengKSMachidaKChengYSLaiMMHepatitis C virus NS3/4A protein interacts with ATM, impairs DNA repair and enhances sensitivity to ionizing radiationVirology2008370229530917931678
- SchogginsJWRiceCMInnate immune responses to hepatitis C virusCurr Top Microbiol Immunol201336921924223463203
- IvanovAVBartoschBSmirnovaOAIsaguliantsMGKochetkovSNHCV and oxidative stress in the liverViruses20135243946923358390
- GongGWarisGTanveerRSiddiquiAHuman hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa BProc Natl Acad Sci U S A200198179599960411481452
- BishéBSyedGSiddiquiAPhosphoinositides in the hepatitis C virus life cycleViruses20124102340235823202467
- ZhangLHongZLinWARF1 and GBF1 generate a PI4P-enriched environment supportive of hepatitis C virus replicationPLoS One201272e3213522359663
- PloenDHafirassouMLHimmelsbachKTIP47 plays a crucial role in the life cycle of hepatitis C virusJ Hepatol20135861081108823354285
- VogtDACamusGHerkerELipid droplet-binding protein TIP47 regulates hepatitis C Virus RNA replication through interaction with the viral NS5A proteinPLoS Pathog201394e100330223593007
- DahariHCotlerSJLaydenTJPerelsonASUnderstanding triphasic HCV decline during treatment in the era of IL28B polymorphisms and direct acting antiviral agents via mathematical modelingJ Hepatol201358484084223246507
- GuedjJDahariHRongLModeling shows that the NS5A inhibitor daclatasvir has two modes of action and yields a shorter estimate of the hepatitis C virus half-lifeProc Natl Acad Sci U S A2013110103991399623431163
- MacdonaldAHarrisMHepatitis C virus NS5A: tales of a promiscuous proteinJ Gen Virol200485Pt 92485250215302943
- MasakiTSuzukiRMurakamiKInteraction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particlesJ Virol200882167964797618524832
- MaYAnantpadmaMTimpeJMHepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteinsJ Virol2011851869720962101
- GouklaniHBeyerCDrummerHGowansEJNetterHJHaqshenasGIdentification of specific regions in hepatitis C virus core, NS2 and NS5A that genetically interact with p7 and co-ordinate infectious virus productionJ Viral Hepat2013204e66e7123490391
- QuezadaEMKaneCMThe stimulatory mechanism of hepatitis C virus NS5A protein on the NS5B catalyzed replication reaction in vitroOpen Biochem J20137111423407362
- QuezadaEMKaneCMThe hepatitis C virus NS5A stimulates NS5B during in vitro RNA synthesis in a template specific mannerOpen Biochem J20093394819590581
- FosterTLBelyaevaTStonehouseNJPearsonARHarrisMAll three domains of the hepatitis C virus nonstructural NS5A protein contribute to RNA bindingJ Virol201084189267927720592076
- Targett-AdamsPGrahamEJMiddletonJSmall molecules targeting hepatitis C virus-encoded NS5A cause subcellular redistribution of their target: insights into compound modes of actionJ Virol201185136353636821507963
- LeeCMaHHangJQThe hepatitis C virus NS5A inhibitor (BMS-790052) alters the subcellular localization of the NS5A non-structural viral proteinVirology20114141101821513964
- CamusGHerkerEModiAADiacylglycerol acyltransferase-1 localizes hepatitis C virus NS5A protein to lipid droplets and enhances NS5A interaction with the viral capsid coreJ Biol Chem2013288149915992323420847
- ReissSRebhanIBackesPRecruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartmentCell Host Microbe201191324521238945
- CarvalhoFACarneiroFAMartinsICDengue virus capsid protein binding to hepatic lipid droplets (LD) is potassium ion dependent and is mediated by LD surface proteinsJ Virol20128642096210822130547
- SamsaMMMondotteJAIglesiasNGDengue virus capsid protein usurps lipid droplets for viral particle formationPLoS Pathog2009510e100063219851456
- LemmJAO’BoyleD2ndLiuMIdentification of hepatitis C virus NS5A inhibitorsJ Virol201084148249119812153
- LemmJALeetJEO’BoyleDRDiscovery of potent hepatitis C virus NS5A inhibitors with dimeric structuresAntimicrob Agents Chemother20115583795380221576451
- GaoMNettlesREBelemaMChemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effectNature201046572949610020410884
- ConteIGiulianoCErcolaniCSynthesis and SAR of piperazinyl-N-phenylbenzamides as inhibitors of hepatitis C virus RNA replication in cell cultureBioorg Med Chem Lett20091961779178319216075
- BarnesMDennisonHMatthewsNSpencerKinventorsArrow Therapeutics Ltd, assigneeMorpholinylanilinoquinazoline derivatives for use as antiviral agents US patent US 20080311076A1
- BiancoAReghellinVDonniciLMetabolism of phosphatidylinositol 4-kinase IIIα-dependent PI4P Is subverted by HCV and is targeted by a 4-anilino quinazoline with antiviral activityPLoS Pathog201283e100257622412376
- LimYSHwangSBHepatitis C virus NS5A protein interacts with phosphatidylinositol 4-kinase type IIIalpha and regulates viral propagationJ Biol Chem201128613112901129821297162
- AmblardFZhangHZhouLSynthesis and evaluation of non-dimeric HCV NS5A inhibitorsBioorg Med Chem Lett20132372031203423466233
- ZhangHZhouLAmblardFSynthesis and evaluation of novel potent HCV NS5A inhibitorsBioorg Med Chem Lett201222144864486822704887
- ShiJZhouLAmblardFSynthesis and biological evaluation of new potent and selective HCV NS5A inhibitorsBioorg Med Chem Lett201222103488349122507961
- ChangWMosleyRTBansalSInhibition of hepatitis C virus NS5A by fluoro-olefin based γ-turn mimeticsBioorg Med Chem Lett20122282938294222425564
- AghemoAColomboMSelection of resistant-associated variants to the NS5A inhibitor daclatasvir: revenge of the hepatitis C virusGastroenterology2013145124724923726875
- FridellRAQiuDWangCValeraLGaoMResistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon systemAntimicrob Agents Chemother20105493641365020585111
- FridellRAWangCSunJHGenotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlationsHepatology20115461924193521809362
- O’Boyle IiDRSunJHNowerPTCharacterizations of HCV NS5A replication complex inhibitorsVirology20134441–234335423896639
- NettlesREGaoMBifanoMMultiple ascending dose study of BMS-790052, a nonstructural protein 5A replication complex inhibitor, in patients infected with hepatitis C virus genotype 1Hepatology20115461956196521837752
- PawlotskyJMNS5A inhibitors in the treatment of hepatitis CJ Hepatol201359237538223567084
- LokASGardinerDFLawitzEPreliminary study of two antiviral agents for hepatitis C genotype 1N Engl J Med2012366321622422256805
- EversonGTSimsKDRodriguez-TorresMInterim analysis of an interferon (IFN)- and ribavirin (RBV))-free regimen of daclatasvir (DCV), asunaprevir (ASV), and BSM-791325 in treatment-naive, hepatitis C virus genotype 1-infected patientsJ Hepatol201358Suppl 1S573 abstract
- WongKAWorthAMartinRCharacterization of hepatitis C virus resistance from a multiple dose clinical trial of the novel NS5A inhibitor GS-5885Antimicrob Agents Chemother Epub7222013
- LawitzEJGruenerDHillJMA phase 1, randomized, placebo-controlled, 3-day, dose-ranging study of GS-5885, an NS5A inhibitor, in patients with genotype 1 hepatitis CJ Hepatol2012571243122314425
- GaneEHylandRDingXELECTRON: 100% suppression of viral load through 4 weeks’ post-treatment for sofosbuvir ledipasvir (gs-5885) ribavirin for 12 weeks in treatment-naïve and -experienced hepatitis C virus GT 1 PatientsPresented at the 20th Conference on Retroviruses and Opportunistic InfectionsMarch 3–6, 2013Atlanta, GA
- Gilead SciencesGilead reports interim data from Phase 2 LONESTAR study [press release]Foster City, CAGilead Sciences2013 [May 2]. Available from: http://investors.gilead.com/phoenix.zhtml?c=69964&p=irol-newsArticle&ID=1814329&highlight#sthash.eUDZEi5S.dpuf2013Accessed October 2, 2013
- MarcellinPMannsMPJanczewskaE12 week response-guided treatment with the NS5A inhibitor, GS-5885, the NS3 protease inhibitor, GS-9451, plus pegylated interferon/ribavirin in treatment naive genotype 1 hepatitis C infected patientsAbstract presented at the 48th Annual Meeting of the European Association for the Study of the LiverApril 24–28, 2013Amsterdam, The Netherlands
- EversonGTdi BisceglieAMVierlingJMCombination of the NS5A inhibitor, GS-5885, the NS3 protease inhibitor, GS-9451, and pegylated interferon plus ribavirin in treatment experienced patients with genotype 1 hepatitis C infectionAbstract presented at the 48th Annual Meeting of the European Association for the Study of the LiverApril 24–28, 2013Amsterdam, The Netherlands
- ThompsonAHanSShiffmanMLGS-5885 GS-9451 Peginterferon and ribavirin (PR) for six or twelve weeks achieves high SVR12 rates in treatment -naïve genotype a IL28B CC patientsAbstract presented at the 48th Annual Meeting of the European Association for the Study of the LiverApril 24–28, 2013Amsterdam, The Netherlands
- Gilead SciencesGilead announces update on Phase 3 study of oral fixed-dose combination of sofosbuvir and ledipasvir for genotype 1 hepatitis C patients [press release]Foster City, CAGilead Sciences2013 [March 26]. Available from: http://investors.gilead.com/phoenix.zhtml?c=69964&p=irol-newsArticle&ID=1800517&highlight#sthash.9dxD7UZn.dpuf2013Accessed October 2, 2013
- KowdleyKVLawitzEPoordadFPhase 2b trial of interferon-free therapy for hepatitis C virus genotype 1N Engl J Med201437022223224428468
- VinceBHillJMLawitzEJA randomized, double-blind, multiple-dose study of the pan-genotypic NS5A inhibitor samatasvir in patients infected with hepatitis C virus genotype 1, 2, 3 or 4J Hepatol Epub2014113
- LahserFLiuRBystolKA combination containing MK5172 (HCV NS3 protease inhibitor) and MK-8742 (HCV NS5A inhibitor) demonstrates high barrier to resistance in HCV repliconPresented at the 63rd Annual Meeting of the American Association for the Study of Liver DiseasesNovember 9–13, 2012Boston, MA
- GaneEFosterGRClanciaraJAntiviral activity, pharmacokinetics, and tolerability of AZD7295, a novel NS5A inhibitor, in a placebo-controlled multiple ascending dose study in HCV genotype 1 and 3 patientsAbstract presented at the 45th Annual Meeting of the European Association for the Study of the LiverApril 14–18, 2010Vienna, Austria
- SpreenWWilfretDBechtelJGSK2336805 HCV NS5A inhibitor demonstrates potent antiviral activity in chronic hepatitis C (CHC) genotype 1 infection: Results from a first time in human (FTIH) single and repeat dose studyPresented at the 62nd Annual Meeting of the American Association for the Study of Liver DiseasesNovember 6–9, 2011San Francisco, CA
- Vertex Pharmaceuticals IncorporatedVertex enters agreement with GlaxoSmithKline for Phase 2 all-oral study of VX-135 and GSK2336805 for the treatment of hepatitis C [press release]Cambridge, MAVertex Pharmaceuticals Incorporated2012 [Nov 1]. Available from: http://investors.vrtx.com/releasedetail.cfm?ReleaseID=7177772013Accessed November 20, 2013
- LalezariJFarrellGShahPPPI-668, a potent new pan-genotypic HCV NS5A inhibitor: Phase 1 efficacy and safetyPresented at the 63rd Annual Meeting of the American Association for the Study of Liver DiseasesNovember 9–13, 2012Boston, MA
- GermanPPangPYangCHealthy volunteer first-in-human evaluation of GS-5816, a novel second generation broad-genotypic NS5A inhibitor with potential for once-daily dosingAbstract presented at the 48th Annual Meeting of the European Association for the Study of the LiverApril 24–28, 2013Amsterdam, The Netherlands
- ChengGYuMPengBGS-5816, a second generation HCV NS5A inhibitor with potent antiviral activity, broad genotypic coverage and a high resistance barrierAbstract presented at the 48th Annual Meeting of the European Association for the Study of the LiverApril 24–28, 2013Amsterdam, The Netherlands
- Gilead SciencesPhase 2 Study of SOF+GS-5816 in treatment naive subjects with chronic HCV Available from: http://clinicaltrials.gov/ct2/show/NCT1858766. NLM identifier: NCT1858766Accessed October 2, 2013
- VinceBLawitzESearleSNovel NS5A inhibitor ACH-2928 Phase I results in HCV GT-1 patientsAbstract presented at the 47th Annual Meeting of the European Association for the Study of the LiverApril 18–22, 2012Barcelona, Spain
- Garcia-RiveraJChatterjiUGallayPCyclophilin inhibitor alisporivir (ALV) combinations with direct acting antivirals reveal strong synergistic anti-HCV effectsAbstract presented at the 48th Annual Meeting of the European Association for the Study of the LiverApril 24–28, 2013Amsterdam, the Netherlands
- LalezariJAgarwalKDusheikoGDose-ranging trial of PPI-461, a potent new pan-genotypic HCV NS5A inhibitor, in patients with HCV genotype-1 infectionAbstract presented at the 62nd Annual Meeting of the American Association for the Study of Liver DiseasesNovember 5–9, 2011San Francisco, CA
- DumasELawalAMenonRPharmacokinetics, safety and tolerability of the HCV NS5a inhibitor ABT-267 following single and multiple doses in healthy adult volunteersAbstract presented at the 46th Annual Meeting of the European Association for the Study of the LiverMarch 30–April 3, 2011Berlin, Germany
- HuiJRobargeLRobisonHKocinskyHSDeshpandeMNo clinically significant pharmacokinetic interaction between sovaprevir and ACH-3102 in healthy volunteersAbstract presented at the 48th Annual Meeting of the European Association for the Study of the LiverApril 24–28, 2013Amsterdam, The Netherlands
- Bristol-Myers SquibbA phase 3 study in combination with BMS-790052 and BMS-650032 in Japanese hepatitis C virus (HCV) patients Available from: http://clinicaltrials.gov/ct2/show/NCT1497834. NLM identifier: NCT1497834Accessed October 10, 2013
- Gilead SciencesSafety and efficacy of sofosbuvir/GS-5885 fixed-dose combination ± ribavirin for the treatment of HCV (ION-2) Available from: http://clinicaltrials.gov/ct2/show/NCT1768286. NLM identifier: NCT1768286Accessed October 2, 2013
- MayersDVinceBHillJIDX719, HCV NS5A inhibitor, demonstrates pan-genotypic activity after three days of monotherapy in genotype 1, 2, 3 or 4 HCV-infected subjectsAbstract presented at the 63rd Annual Meeting of the American Association for the Study of Liver DiseasesNovember 9–13, 2012Boston, MA
- ZhouXJVinceBHillJPharmacokinetics and pharmacodynamics of IDX719, a pan-genotypic HCV NS5A inhibitor in genotype 1, 2, 3 or 4 HCV-infected subjectsAbstract presented at the Asian Pacific Association for the Study of the LiverJune 6–10, 2013Singapore
- MerckSafety, Pharmacokinetics and pharmacodynamics of MK-8742 in hepatitis C infected males (MK-8742–8002 AM1) Available from: http://clinicaltrials.gov/ct2/show/NCT1532973. NLM identifier: NCT1532973Accessed October 10, 2013
- Abbott LaboratoriesAbbott presents promising Phase 2b interferon-free hepatitis C results at 2012 liver meeting® [press release]Abbott Park, ILAbbott Laboratories2012 [November 10]. Available from: http://www.abbott.com/news-media/press-releases/abbott-presents-promising-phase-2b-interferonfree-hepatitis-c-results-at-2012-liver-meeting.htmAccessed June 27, 2013
- clinical trialfacts.com [homepage on the Internet]Effect of omeprazole and ritonavir on GSK2336805 pharmacokinetics in healthy adultsClinicalTrialfacts.com2011 [updated October 1, 2013]. Available from: http://www.clinicaltrialfacts.com/hepatitis-c-chronic/effect-of-omeprazole-and-ritonavir-on-gs-115217/summaryAccessed October 13, 2013
- investorshub.advfn.com [homepage on the Internet]Post 94863Investors Hub2010 Available from: http://investorshub.advfn.com/boards/read_msg.aspx?message_id=49610323Accessed October 13, 2013
- bms.com [homepage on the Internet]Pipeline asset update for daclatasvir (DCV; BMS-790052)Bristol-Myers Squibb2012 [cited July 15, 2013]. Available from: http://www.bms.com/research/investigational/hepatitis/Pages/DCV-BMS-790052.aspxAccessed October 13, 2013

