Abstract
Polyethylenimine (PEI), especially PEI 25 kDa, has been widely studied for delivery of nucleic acid drugs both in vitro and in vivo. However, it lacks degradable linkages and is too toxic for therapeutic applications. Hence, low-molecular-weight PEI has been explored as an alternative to PEI 25 kDa. To reduce cytotoxicity and increase transfection efficiency, we designed and synthesized a novel small-molecular-weight PEI derivative (PEI-Et, Mn: 1220, Mw: 2895) with ethylene biscarbamate linkages. PEI-Et carried the ability to condense plasmid DNA (pDNA) into nanoparticles. Gel retardation assay showed complete condensation of pDNA at w/w ratios that exceeded three. The particle size of polymer/pDNA complexes was between 130 nm and 180 nm and zeta potential was 5–10 mV, which were appropriate for cell endocytosis. The morphology of PEI-Et/pDNA complexes observed by atomic force microscopy (AFM) was spherically shaped with diameters of 110–190 nm. The transfection efficiency of polymer/pDNA complexes as determined with the luciferase activity assay as well as fluorescence-activated cell-sorting analysis (FACS) was higher than commercially available PEI 25 kDa and Lipofectamine 2000 in various cell lines. Also, the polymer exhibited significantly lower cytotoxicity compared to PEI 25 kDa at the same concentration in three cell lines. Therefore, our results indicated that the PEI-Et would be a promising candidate for safe and efficient gene delivery in gene therapy.
Introduction
Gene therapy, the introduction of an extraneous gene into the target cells or organs with the aim of replacing a lost cellular function or introducing a new functionality, offers tremendous promise for the treatment of many inherited and acquired diseases. An efficient and safe delivery system that delivers the therapeutic gene to a specific target tissue or organ is the prerequisite for successful gene therapy.Citation1,Citation2 Gene delivery vectors are classified into two categories: viral and nonviral vectors. Although with lower efficiency when compared with viral vectors,Citation3 nonviral alternatives have been developed as safer strategies for gene delivery owing to several advantages over viral vectors, including safety, lower immunogenicity, the ease of chemical modification and the ability to transfer larger plasmid DNA (pDNA) molecules.Citation4–Citation6 Nonviral vectors can be divided into two categories: cationic polymers and cationic lipid carriers. Cationic polymer/pDNA complexes tend to be more stable than cationic lipid/pDNA complexes.Citation7
Recently, the cationic polyethylenimine (PEI) has been widely used for nonviral transfection. PEI effectively condenses pDNA into nanoparticles and holds capacity for transfecting a variety of cell lines both in vitro and in vivo by force of a proton sponge effect, which prevents the pDNA from endosomal disruption.Citation8,Citation9 The molecular weight of PEI is a critical factor influencing the toxicity and transfection efficiency.Citation10 Low-molecular-weight PEI (<2 kDa) was proven to be nontoxic, ineffectively condensing pDNA, and displaying very poor transfection activity. In contrast, PEI with higher molecular weight (25 kDa) exhibited higher transfection efficiency and yet higher toxicity than other low-molecular-weight PEIs.Citation11,Citation12 In addition, PEI 25 kDa with the structure containing C–C or C–N bonds was totally nonbiodegradable, which would give rise to accumulative cytotoxicity in vivo. Taken together, the long-term safety of high-molecular weight PEI is further problematic. Therefore, to address the conflict, several modified PEI such as PEGylation,Citation13,Citation14 acylationCitation15 of high molecular weight and cross-linked low-molecular-weight PEIs with biodegradable linkages (ester,Citation16,Citation17 disulfide,Citation18 and amide,Citation19 linkages, etc) have been investigated as gene carriers to increase its gene delivery efficiency and reduce cytotoxicity. Recently, Zhao and colleaguesCitation20 synthesized biodegradable PEI–triethylene glycol (TEG) based on PEI 2 kDa and TEG through biscarbamate linkages. They reported that the polymer could be degraded in neutral environment and exhibited high transfection ability with low toxicity compared to PEI 25 kDa. Previously, we also introduced butyl biscarbamate cross-linked PEI derivative (PEIC) with small molecular weight, which displayed higher transfection efficiency and lower cytotoxicity compared to PEI 25 kDa in COS-7 cells. PEIC exhibited the highest gene transfection efficiency when the PEIC/pDNA weight ratio (w/w) was 40.Citation21
In this paper, we designed and synthesized a novel cross-linked PEI derivative with ethylene biscarbamate linkage (PEI-Et). PEI-Et was synthesized and characterized with proton nuclear magnetic resonance (1H-NMR), Fourier transform-infrared (FT-IR), and gel permeation chromatography (GPC). Then PEI-Et/pDNA complexes were prepared and characterized in terms of particle size, zeta potential, pDNA condensation ability, and morphology under atomic force microscope (AFM). In addition, the cytotoxicities of the polymer were determined with cell viability assay, the transfection efficiencies of PEI-Et/pDNA complexes were evaluated in several cell lines including COS-7, HeLa, and BRL-3A cells with luciferase activity assay as well as fluorescence microscopy and fluorescence-activated cell-sorting analysis (FACS). PEI-Et also had small molecular weight compared with PEI 25 kDa. In comparison to PEIC’s best transfection performance at PEIC/pDNA w/w of 40 in COS-7 cell lines, PEI-Et/pDNA complexes exhibited best transfection activity with w/w ranging 10–20 in different cell lines, which would favor its further functional gene investigation due to the decreased w/w ratio. In addition, PEI-Et displayed lower toxicity than PEIC in COS-7 and BRL-3A cell lines (data not shown). Our results demonstrated that PEI-Et had the potential to be a promising candidate as a nonviral gene carrier.
Materials and methods
Materials
Branched PEI (25 kDa, 800 Da), ethylene bis(chloroformate), ethidium bromide (EB), and 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazoliumbromide (MTT) were purchased from Sigma-Aldrich (St Louis, MO). MicroBCA protein assay kit was obtained from Pierce (Rockford, IL). Luciferase assay kit was purchased from Promega (Madison, WI). All other chemicals were of analytical grade.
Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS) and Trypsin-EDTA were obtained from PAA (Cölbe, Germany). The plasmids used in the study were pGL3-Control (Promega) encoding firefly luciferase and pEGFP-N1 (Clontech, Palo Alto, CA) encoding enhanced green fluorescent protein (EGFP). The purity of the plasmids was checked by electrophoresis on a 1% agarose gel, and the concentration of pDNA was determined by spectrophotometry (A260/A280).
Cell culture
The cell lines COS-7 (African green monkey kidney fibroblast) BRL-3A (normal rat liver cell), and HeLa (human cervix epithelial carcinoma cell) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM medium supplemented with 10% FBS. Cells were maintained at 37°C in a humidified 5% CO2 incubator.
Synthesis of PEI-Et
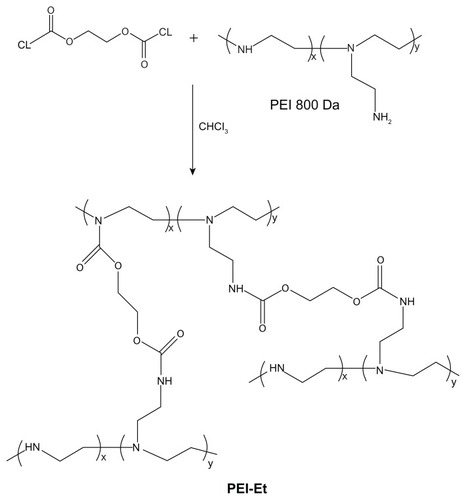
Branched PEI, 800 Da in average molecular weight (Mw), and ethylene bis(chloroformate) were respectively dissolved in freshly anhydrous chloroform. The ethylene bis(chloroformate) solution (10 mg/mL, 18.7 mL) was added dropwise to a PEI solution (300 mg/mL, 3 mL) with stirring. The reaction was carried out in an ice bath in a nitrogen atmosphere and allowed to proceed overnight. Then the sample was evaporated under reduced pressure to remove the solvent and vacuum-dried for 24 hours. The sample was subsequently dissolved in water and dialyzed against distilled water in a dialysis tube (MWCO: 1000 Da) for 2 days and lyophilized to yield the polymer. The polymer was stored at −20°C for further use.
Characterization of polymer
PEI-Et was estimated by measuring proton nuclear magnetic resonance (1H-NMR) (Mercury Plus 400; Varian Inc, Palo Alto, CA) and FT-IR. 1H-NMR spectra was recorded on a Varian Unity 300 MHz spectrometer (Varian Inc) using D2O as a solvent. The FT-IR spectrum of polymer was recorded in pressed KBr pellets on a FT-IR spectrometer (Equinox 55; Bruker Optics, Bruck, Germany). The molecular weight of the polymer was measured by GPC relative to PEG standards (Mw range Mp: 106, 430, 633, 1400, 4290, 7130, 12,600, 20,600 Da), using a Waters high-pressure liquid chromatography (HPLC) system (Milford, MA) equipped with a gel permeation column (GPC) and a refractive index detector. The HPLC mobile phase was ultrapure water.
Preparation of PEI-Et/pDNA complexes
PEI-Et/pDNA complexes were freshly prepared prior to use. Briefly, both PEI-Et and pDNA were separately diluted to the appropriate concentration in phosphate-buffered saline (PBS; pH 7.4, mimicking pH in physiological condition) depending on the required w/w ratio, then the complexes were prepared by adding polymer solutions to equal volumes of pDNA with gentle vortexing and incubated at room temperature for 30 minutes before use.
Gel retardation assay
The pDNA condensation ability of the polymer was evaluated by agarose gel electrophoresis with plasmid of enhanced green fluorescent protein (pEGFP). PEI-Et/pDNA and PEI 800 Da/pDNA complexes were prepared at various w/w ratios from 1 to 30. After incubation for 30 minutes at room temperature, the complexes were mixed with appropriate amounts of 5 × gel-loading buffer and then electrophoresed on 1% (w/v) agarose gels containing EB (0.5 μg/mL of the gel) with 1 × Tris-acetate (TAE) buffer at 80 V for 40 minutes. The locations of the pDNA bands were visualized with a UV illuminator (Fisher Biotec, Australia).
Particle size and zeta potential measurements
The particle size and zeta potential of complexes were measured using a particle size analyzer (90Plus; Brookhaven, Holtsville, NY). PEI-Et/pDNA and PEI 800 Da/pDNA complexes were prepared at various w/w ratios from 0.5 to 30. The measurement was performed at 25°C with a scattering angle of 90° and an electric field strength of 7 V/cm. Each sample was performed in triplicate.
Atomic force microscopy
The morphology of complexes at w/w ratio 20 was observed by atomic force microscopy (AFM). One drop of the complexes was placed on a mica disk, and dried at room temperature. Then it was examined under AFM (E-Sweep; SII Nanotechnology, Inc, Chiba, Japan). The images were taken in air under tapping mode with standard 125-mm single-crystal silicon cantilevers.
Cytotoxicity assay
MTT assay was employed to measure the cytotoxicity of the polymer, with PEI 25 kDa as a control. Cells were seeded in 96-well plates at an initial density of 5000 cells/well in 100 μL of DMEM containing 10% FBS. After incubation for 24 hours, the media were replaced with fresh serum-free DMEM containing polymers at various concentrations or polymer/pDNA complexes at various w/w ratios (2, 5, 10, 20, 30, 50). After an additional incubation for 4 hours, the media were changed with fresh serum-free DMEM and 25 μL of stock solution of MTT (5 mg/mL in PBS) was added to each well. The cells were incubated for a further 6 hours, afterwards, the media were removed and 150 μL of dimethyl sulfoxide (DMSO) was added to the well, the plates were mildly shaken for 15 minutes to ensure the dissolution of formazan. Finally, the absorbance was measured using an ELISA reader (MK3; Thermo Labsystem, Vantaa, Finland) at 570 nm (with 630 nm as a reference wavelength). The cell viability was expressed as a percentage of the absorbance to that of the control experiment without polymers. Five replicates were counted for each sample.
Transfection experiments in vitro
Luciferase activity assay
PEI-Et-mediated transfections were evaluated in COS-7, BRL-3A, and HeLa cells by using the reporter plasmid pGL3- control. Cells were seeded in 48-well plates at an initial density of 5 × 104 cells/well in 500 mL of DMEM containing 10% FBS. After incubation for 24 hours, The cells were washed with PBS followed by the addition of serum-free or serum-containing DMEM with polymer/pGL3-control (500 ng) complexes at various w/w ratios and additionally incubated for 4 hours. Cells transfected with naked pDNA were used as negative control, cells transfected with PEI 25 kDa/pDNA (optimal w/w), Lipofectamine 2000/pDNA (optimal w/w), and PEI 800 Da/pDNA were used as positive controls. Afterwards, the media were exchanged for 500 mL of fresh DMEM containing 10% FBS and allowed to incubate for 44 hours. Then the cells were washed with PBS before adding 75 μL of cell lysis buffer, and incubated for 30 minutes. The luciferase assay was carried out according to the manufacturer’s instructions (Promega). The relative light units (RLU) of luciferase expression were measured with a luminometer. The protein concentration in the cell lysates was measured by MicroBCA protein assay kit. The luciferase activity was expressed in terms of RLU/mg protein. Each sample was performed in triplicate.
Fluorescence microscopy and fluorescence-activated cell-sorting analysis
The optimal w/w ratio of PEI-Et/pEGFP-N1 complexes in HeLa cells as determined from the luciferase activity assay was selected for this experiment. The levels of green fluorescent protein (GFP) expression were characterized using fluorescence microscopy (Olympus, Tokyo, Japan). The transfection efficiency was quantified by the percentage of cells expressing GFP and analyzed by flow cytometry using a FACS Caliber System from Becton-Dickinson (San Jose, CA). The data was calculated based on measurements from three individual experiments.
Statistical analysis
All statistical analyses were performed with SPSS software (v 19.0; SPSS Inc, Chicago, IL). Data were illustrated as mean ± standard deviation. Student’s t-test (two-tailed) was applied to test the significance of the differences between two groups. Data were considered significant difference at the level of P < 0.05 and very significant difference at the level of P < 0.01.
Results and discussion
Synthesis and characterization of polymer
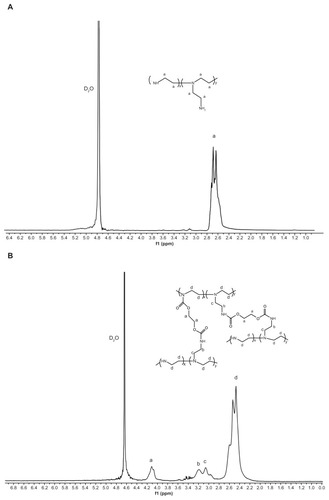
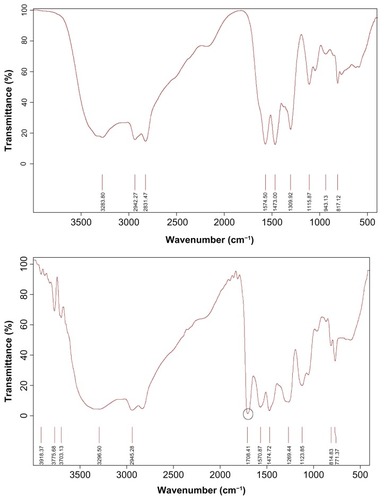
PEI-Et was synthesized through the conjugation of ethylene bis(chloroformate) to PEI 800 Da (). The structure of PEI-Et was confirmed using 1H-NMR and FT-IR. In , peak a was from the methylene protons of (–CH2CH2 NH–) in PEI 800 Da. As for , peak a was attributed to the protons of (–OCOCH2CH2OCO–), peaks b and c were from the methylene protons of (–NH–CH2CH2 NH–CO–) in the side chain of PEI-Et, and peak d was assigned to the methylene protons of (–CH2CH2 NH–) in the backbone. In the FT-IR spectrum (), the stretching vibration at 1708 cm−1 appeared and was attributed to the carbamate carbonyl group of PEI-Et. The average molecular weight of PEI-Et measured with a GPC instrument was 2895 Da, with a polydispersity of 2.37. These results demonstrated that ethylene bis(chloroformate) was successfully cross-linked to PEI 800 Da.
Figure 1 Reaction scheme of PEI-Et.
Abbreviations: PEI, polyethylenimine; PEI-Et, PEI derivative with ethylene biscarbamate linkage.
Characterization of PEI-Et/pDNA complexes
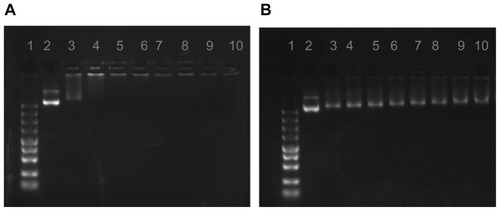
The condensation of pDNA into small particles is a necessary prerequisite for gene delivery using cationic polymers.Citation22 The formation of PEI-Et/pDNA complexes was based on electrostatic interaction between the positive amino groups of PEI-Et and negative phosphate groups of pDNA. The pDNA condensation ability of the polymer was confirmed by agarose gel electrophoresis. As shown in , the migration of pDNA was completely retarded when w/w ratio reached approximately three, indicating that pDNA was completely complexed with PEI-Et at w/w ratios ≥ 3. Interaction of DNA or small interfering RNA (siRNA) with cationic polymers could protect the condensed pDNA or siRNA from enzymatic degradation,Citation23,Citation24 making it more effective for cellular transfection. However, PEI 800 Da as the backbone of PEI-Et could not retard the migration of pDNA even at a w/w ratio of 30 (), indicating that PEI 800 Da was incapable of forming complexes with pDNA.
Figure 4 Agarose gel electrophoresis of PEI-Et/pDNA (A) and PEI 800 Da/pDNA (B) complexes at various w/w ratios.
Notes: Lane 1, Marker; Lane 2, naked pDNA; Lanes 3–10: polymer/pDNA complexes at w/w ratios of 1, 3, 5, 7, 10, 15, 20, 30.
Abbreviations: pDNA, plasmid DNA; PEI, polyethylenimine; PEI-Et, PEI derivative with ethylene biscarbamate linkage.
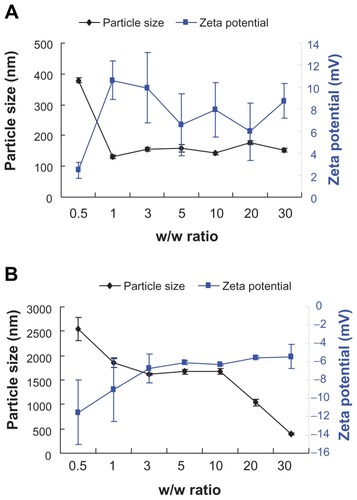
The particle size of the polymer/pDNA complexes was a critical factor for gene carriers and could affect the transfection efficiency.Citation25 A positive surface charge of untargeted polyplexes was necessary for binding to the negatively charged cellular membrane, which in turn facilitated uptake into the cell.Citation26–Citation28 showed that at a w/w ratio of 0.5, the particle size of PEI-Et/pDNA complexes was 379 nm, the zeta potential was 2 mV, indicating poor complexation between pDNA and PEI-Et. However, with an increase in w/w ratio from 1 to 30, PEI-Et could efficiently condense pDNA into nanoparticles with relatively constant diameters ranging from 130 nm to 180 nm, suggesting the formation of stable complexes with a size appropriate for gene delivery. At the same time, zeta potential ranged from 5 mV to 10 mV. In the case of PEI 800 Da (), the particle size decreased drastically with increasing w/w ratio. PEI 800 Da formed loose complexes with a size of approximately 400 nm even at w/w 30. The zeta potential was negative at w/w from 0.5 to 30. These results also confirmed the results of the gel retardation assay.
Figure 5 Particle size and zeta potential of PEI-Et/pDNA (A) and PEI 800 Da/pDNA (B) complexes as determined by dynamic light scattering at various w/w ratios.
Note: n = 3, error bars represent standard deviation.
Abbreviations: pDNA, plasmid DNA; PEI, polyethylenimine; PEI-Et, PEI derivative with ethylene biscarbamate linkage.
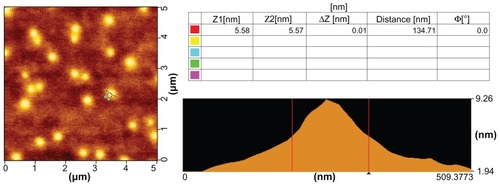
showed representative morphologies of PEI-Et/pDNA complexes under AFM. The result indicated that the complexes were spherically shaped with diameters of 110–190 nm, which was in agreement with the particle size using dynamic light scattering.
Cytotoxicity assay
Cytotoxicity was a major hurdle for clinical feasibility of polycationic gene carriers.Citation29 There were at least two types of cytotoxicity mediated by PEI-Et: an immediate toxicity associated with free PEI-Et and a delayed toxicity associated with PEI-Et/pDNA complexes. In this study, free polymers as well as polymer/pDNA complexes were selected to measure the cytotoxicity. Free polymers were used to simulate a worst case scenario and obtain larger sensitivity results, since toxicity was reduced when polymer/pDNA complexes were formed.Citation26 Cytotoxicity profiles were analyzed by MTT assay in COS-7, BRL-3A, and HeLa cell lines.
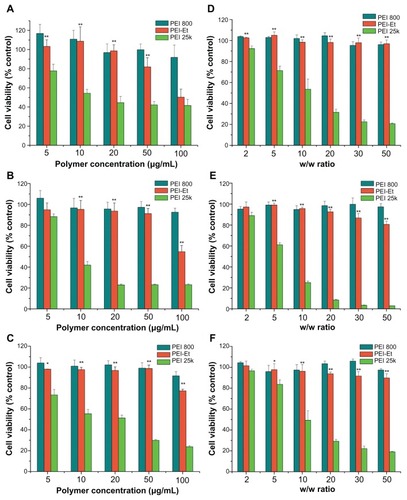
indicated that PEI 800 Da and PEI 800 Da/pDNA complexes showed very low cytotoxicity. The cell viabilities were over 91% at the tested concentrations or w/w ratios. PEI-Et exhibited much lower cytotoxicity than PEI 25 kDa at the same concentration in three different cell lines. In addition, PEI-Et produced almost no cytotoxicity at concentrations below 50 μg/mL. The cell viabilities were 103% ± 7%, 94% ± 6%, and 98% ± 1% at a polymer concentration of 5 μg/mL in COS-7, BRL-3A, and HeLa cells, respectively. And these values slightly decreased to 81% ± 9%, 91% ± 4%, and 98% ± 3%, respectively with the polymer concentration increasing to 50 μg/mL, which implied that a wide dose range of PEI-Et may be used for gene delivery. However, the cell viability attenuated drastically when the concentrations exceeded 50 μg/mL, exhibiting a concentration-dependent property. In contrast, cell viability decreased remarkably with increasing concentrations of PEI 25 kDa. For example, the cell viabilities were from 77% ± 6%, 88% ± 2%, and 73% ± 5% at a PEI 25 kDa concentration of 5 μg/mL to 41% ± 6%, 23.11% ± 1%, and 23.71% ± 1% at a PEI 25 kDa concentration of 100 μg/mL in COS-7, BRL-3A, and HeLa cells, respectively. For the cell viabilities of polymer/pDNA complexes, PEI-Et/pDNA complexes also displayed remarkably low cytotoxicity than PEI 25 kDa/pDNA complexes among the three cell lines.
Figure 7 Cytotoxicity of the polymers at various concentrations and cytotoxicity of the polymer/pDNA complexes at various w/w ratios in COS-7 (A and D), BRL-3A (B and E), and HeLa (C and F) cell lines.
Notes: n = 5, error bars represent standard deviation, *P < 0.05, **P < 0.01 vs PEI 25 kDa.
Abbreviations: pDNA, plasmid DNA; PEI, polyethylenimine; PEI-Et, PEI derivative with ethylene biscarbamate linkage.
Generally, the cytotoxicity of cationic polymer was a consequence of polymer aggregation on the cell surface due to strong electrostatic interactions in the plasma membrane, which impaired cell membrane function.Citation11,Citation30 The molecular weight was a major factor affecting the cytotoxicity of PEI, low-molecular-weight PEI produced lower cytotoxicity when compared with high-molecular-weight PEI.Citation12 For this reason, PEI-Et with much lower cytotoxicity was probably due to the significantly lower molecular weight than PEI 25 kDa. This was in agreement with a study Bieber and ElsasserCitation31 previously reported, Bieber separated high-molecular-weight PEI into fractions of different molecular weight by gel filtration chromatography (GFC) and found a positive correlation between cytotoxicity and molecular weight. Our results demonstrated that PEI-Et was a very promising material for safe gene delivery compared to PEI 25 kDa.
Transfection experiments in vitro
Luciferase activity assay
To investigate the in vitro gene delivery efficiency of PEI-Et, we performed luciferase activity assays, using three different cell lines described above. PEI 800 Da, PEI 25 kDa, and Lipofectamine 2000 at optimal w/w were used as positive controls. As illustrated in , transfection efficiency of the polymer was dependent on the cell line, and the highest transfection efficiency was obtained in HeLa cell line. Naked pDNA yielded almost negligible luciferase expression, implying that pDNA without any carrier exhibited fairly low transfection efficiency according to the study reported previously.Citation32,Citation33 Also, interestingly, transfection efficiency of PEI-Et increased with increasing w/w ratios below 20, and then decreased at higher w/w ratios in all the three cell lines. This phenomenon could be explained that a low w/w ratio would yield physically unstable complexes and poor transfection, whereas a high w/w ratio resulted in a poor transfection due to their stability, thus, the pDNA could not be released from the complexes.
Figure 8 Transfection efficiency of PEI-Et/pGL3-Control complexes at various w/w ratios in serum-free medium in (A) COS-7, (B) BRL-3A, and (C) HeLa cell lines, and (D) transfection efficiency of PEI-Et/pGL3-Control complexes (w/w 20) in serum-containing medium in the three cell lines, in comparison with that of PEI 25 kDa (w/w 2), Lipofectamine 2000 (w/w 1), and PEI 800 Da (w/w 2).
Notes: n = 3, error bars represent standard deviation, *P < 0.05, **P < 0.01.
Abbreviations: N, naked pDNA; 25k, PEI 25 kDa; L, Lipofectamine 2000; PEI, polyethylenimine; PEI-Et, PEI derivative with ethylene biscarbamate linkage.
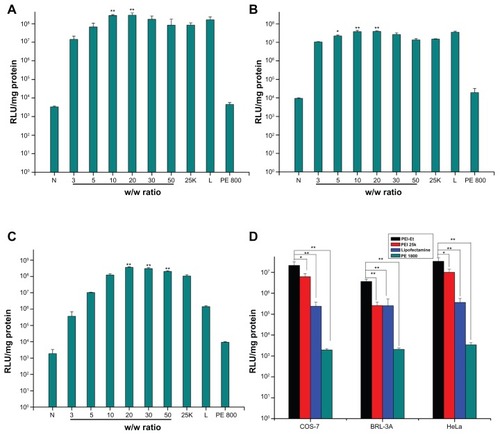
In addition, PEI-Et/pDNA complexes at w/w ratio 20 showed a 3.4-fold higher transfection efficiency in comparison to PEI 25 kDa at its optimal w/w ratio 2 in COS-7 cells (P < 0.01). In BRL-3A cells, PEI-Et/pDNA complexes at w/w 20 resulted in a 2.6-fold higher transfection efficiency in comparison to PEI 25 kDa at its optimal w/w 2 (P < 0.01). In HeLa cells, PEI-Et/pDNA complexes at w/w 20 resulted in a 3.5-fold higher transfection efficiency than PEI 25 kDa at its optimal w/w 2 (P < 0.01), and 25-fold higher transfection efficiency than Lipofectamine 2000 at its optimal w/w ratio 1 (P < 0.01). No statistically significant differences existed between the data of complexes at w/w ratio 20 and the data of the Lipofectamine 2000 at its optimal w/w ratio 1 in both COS-7 and BRL-3A cells (P > 0.05). Transfection was also performed in serum-containing medium, and w/w 20 for PEI-Et/pDNA complexes that produced the highest transfection efficiency in serum-free condition was selected. As shown in , for all the polymers, the presence of serum inhibited the transfection activity. This was possibly due to the binding of the positively charged polymer/pDNA complexes with the negatively charged serum proteins that would reduce the cellular uptake of complexes.Citation34 However, PEI-Et/pDNA complexes still exhibited much higher transfection efficiency than PEI 25 kDa (P < 0.05), Lipofectamine 2000 (P < 0.01) and PEI 800 Da (P < 0.01) in the three cell lines. The enhanced transfection efficiency of PEI-Et than PEI 25 kDa was probably due to the weaker condensation capability of PEI-Et than PEI 25 kDa, which would lead to an easy release of pDNA from the complexes after passing through the cell membrane. Moreover, the reduced cytotoxicity would also facilitate the transfection efficiency. These results indicated that the polymer was more efficient as a gene carrier, compared with the commercial transfection reagent PEI 25 kDa, which was considered as the most popular “gold standard” in gene transfection, and was comparable to Lipofectamine 2000.
Fluorescence microscopy and fluorescence-activated cell-sorting analysis
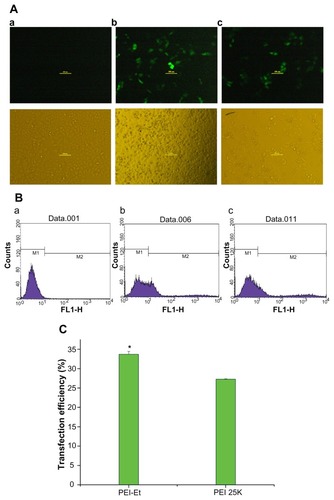
To confirm the enhanced transfection efficiency of PEI-Et than commercially available PEI 25 kDa, gene delivery efficiency was evaluated in HeLa cells, using pEGFP-N1 as a reporter gene. PEI-Et/pDNA (w/w 20) and PEI 25 kDa/pDNA (w/w 2) were selected based on the luciferase activity assay. In the fluorescence microscope images (), the cells transfected with PEI-Et/pEGFP-N1 displayed more bright green fluorescent spots than PEI 25 kDa/pEGFP-N1. Furthermore, the transfection efficiency was quantified using flow cytometry in terms of cell population expressing GFP. As shown in , transfection efficiency of PEI-Et/pDNA (33%) was higher than that of PEI 25 kDa/pDNA (27%), with P < 0.05. These results accorded well with the luciferase activity assays, indicating the superiority of PEI-Et over PEI 25 kDa.
Figure 9 Fluorescence images (A) and flow cytometry analyzed graphs (B) of cell distribution expressing GFP of HeLa cells transfected with (a) PBS (negative control), (b) PEI-Et/pEGFP-N1 (w/w 20), (c) PEI 25 kDa/pEGFP-N1 (w/w 2) (magnification, 100× ), and (C) transfection efficiency of PEI-Et and PEI 25 kDa as a percentage of EGFP positive cells per total amount of HeLa cells.
Notes: n = 3, error bars represent standard deviation, *P < 0.05.
Abbreviations: PEI, polyethylenimine; PEI-Et, PEI derivative with ethylene biscarbamate linkage.
Conclusion
In the present study, a novel PEI derivative PEI-Et linked through biscarbamate linkages was successfully prepared and evaluated as nonviral gene carriers. The PEI-Et was synthesized by a simple chemical procedure, carried the enhanced ability to condense pDNA into nanoparticles and physicochemical properties appropriate for gene delivery. The polymer exhibited much lower cytotoxicity and significantly enhanced transfection efficiency in various cell lines when compared to commercially available PEI 25 kDa. Therefore, it is reasonable to conclude that PEI-Et would be a promising strategy for safe and efficient gene delivery in gene therapy.
Acknowledgments
This work was supported by grants from the program of major scientific and technological specialized project for “significant new formulation of new drugs” (2009ZX09310-007), the National Natural Science Foundation of China (81001416), and the Shanghai Science and Technology Committee, China (10JC1408902, 1052nm3900, and 0952nm03700). We also thank the Analytical Center of Shanghai Jiao Tong University for technical support.
Disclosure
The authors report no conflicts of interest in this work.
References
- ParkTGJeongJHKimSWCurrent status of polymeric gene delivery systemsAdv Drug Deliv Rev20065846748616781003
- SrinivasRSamantaSChaudhuriACationic amphiphiles: promising carriers of genetic materials in gene therapyChem Soc Rev2009383326333820449052
- DittoAJShahPNYunYHNon-viral gene delivery using nanoparticlesExpert Opin Drug Deliv200961149116019780712
- MintzerMASimanekEENonviral vectors for gene deliveryChem Rev200910925930219053809
- ThomasMKlibanovAMNon-viral gene therapy: polycation-mediated DNA deliveryAppl Microbiol Biotechnol200362273412719940
- YuHWagnerEBioresponsive polymers for nonviral gene deliveryCurr Opin Mol Ther20091116517819330722
- ParkDWHoffmanASPunSDesign and development of polymers for gene deliveryNat Rev Drug Discov2005458159316052241
- DemeneixBBehrJBoussifOGene transfer with lipospermines and polyethyleniminesAdv Drug Deliv Rev199830859510837604
- BoussifOLezoualc’hFZantaMAA versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimineProc Natl Acad Sci U S A199592729773017638184
- GodbeyWTWuKKMikosAGSize matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicleJ Biomed Mater Res A199945268275
- FischerDBieberTLiYA novel non-viral vector for DNA delivery based on low molecular weight, branched polyethylenimine: effect of molecular weight on transfection efficiency and cytotoxicityPharm Res1999161273127910468031
- GraysonACDoodyAMPutnamDBiophysical and structural characterization of polyethylenimine-mediated siRNA delivery in vitroPharm Res2006231868187616845585
- LutzGJSirsiSRWilliamsJHPEG-PEI copolymers for oligonucleotide delivery to cells and tissuesMethods Mol Biol200843314115818679622
- ZhangXPanSRHuHMPoly(ethylene glycol)-block-polyethylenimine copolymers as carriers for gene delivery: effects of PEG molecular weight and PEGylation degreeJ Biomed Mater Res A20088479580417635020
- ForrestMLMeisterGEKoerberJTPartial acetylation of polyethylenimine enhances in vitro gene deliveryPharm Res20042136537115032320
- GuoQFLiuTTYanXSynthesis and properties of a novel biodegradable poly(ester amine) copolymer based on poly(l-lactide) and low molecular weight polyethylenimine for gene deliveryInt J Nanomedicine201161641164921904454
- ThomasMGeQLuJJCross-linked small polyethylenimines: while still nontoxic, deliver DNA efficiently to mammalian cells in vitro and in vivoPharm Res20052237338015835742
- PengQZhongZZhuoRDisulfide cross-linked polyethylenimines (PEI) prepared via thiolation of low molecular weight PEI as highly efficient gene vectorsBioconjug Chem20081949950618205328
- KloecknerJWagnerEOgrisMDegradable gene carriers based on oligomerized polyaminesEur J Pharm Sci20062941442517000090
- ZhaoDGongTZhuDComprehensive comparison of two new biodegradable gene carriersInt J Pharm201141326027021536113
- XuSLChenMYYaoYNovel poly(ethylene imine) biscarbamate conjugate as an efficient and nontoxic gene delivery systemJ Control Release2008130646818582980
- NeuMFischerDKisselTRecent advances in rational gene transfer vector design based on poly(ethylene imine) and its derivativesJ Gene Med20057992100915920783
- HaradaATogawaHKataokaKPhysicochemical properties and nuclease resistance of antisense-oligonucleotides entrapped in the core of polyion complex micelles composed of poly(ethylene glycol)-poly(l-lysine) block copolymersEur J Pharm Sci200113354211292566
- JeongJHKimSWParkTGNovel intracellular delivery system of antisense oligonucleotide by self assembled hybrid micelles composed of DNA/PEG conjugate and cationic fusogenic peptideBioconjug Chem20031447347912643759
- JiangHLKimYKAroteRChitosan-graft-polyethylenimine as a gene carrierJ Control Release200711727328017166614
- KunathKvon HarpeAFischerDLow-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimineJ Control Release20038911312512695067
- ParkMRHanKOHanIKDegradable polyethylenimine-alt-poly(ethylene glycol) copolymers as novel gene carriersJ Control Release200510536738015936108
- ZhangXQIntraJSalemAKConjugation of polyamidoamine dendrimers on biodegradable microparticles for nonviral gene deliveryBioconjug Chem2007182068207617848077
- LvHTZhangSBWangBToxicity of cationic lipids and cationic polymers in gene deliveryJ Control Release200611410010916831482
- RyserHJA membrane effect of basic polymers dependent on molecular sizeNature19672159349366055419
- BieberTElsasserHPPreparation of a low molecular weight polyethylenimine for efficient cell transfectionBiotechniques200130748111196323
- GaoYZhangZWChenLLSynthesis of 6-N,N,N-trimethyltriazole chitosan via “click chemistry” and evaluation for gene deliveryBiomacromolecules2009102175218219722556
- GaoYXuZChenSArginine–chitosan/DNA self-assemble nanoparticles for gene delivery: in vitro characteristics and transfection efficiencyInt J Pharm200835924124618479851
- YangJPHuangLOvercoming the inhibitory effect of serum on lipofection by increasing the charge ratio of cationic liposome to DNAGene Ther199749509609349432