 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
MicroRNAs are small (18–22 nucleotide long) noncoding RNAs that play important roles in biological processes through posttranscriptional regulation of gene expression. Their aberrant expression and functional significance are reported in several human malignancies, including pancreatic cancer. Recently, we identified miR-150 as a novel tumor suppressor microRNA in pancreatic cancer. Furthermore, expression of miR-150 was downregulated in the majority of tumor cases, suggesting that its restoration could serve as an effective approach for pancreatic cancer therapy. In the present study, we developed a nanoparticle-based miR-150 delivery system and tested its therapeutic efficacy in vitro. Using double emulsion solvent evaporation method, we developed a poly (D,L-lactide-co-glycolide) (PLGA)-based nanoformulation of miR-150 (miR-150-NF). Polyethyleneimine (a cationic polymer) was incorporated in PLGA matrix to increase the encapsulation of miR-150. Physical characterization of miR-150-NF demonstrated that these nanoparticles had high encapsulation efficiency (~78%) and exhibited sustained release profile. Treatment of pancreatic cancer cells with miR-150-NF led to efficient intracellular delivery of miR-150 mimics and caused significant downregulation of its target gene (MUC4) expression. Inhibition of MUC4 correlated with a concomitant decrease in the expression of its interacting partner, HER2, and repression of its downstream signaling. Furthermore, treatment of pancreatic cancer cells with miR-150-NF suppressed their growth, clonogenicity, motility, and invasion. Together, these findings suggest that PLGA-based nanoformulation could potentially serve as a safe and effective nanovector platform for miR-150 delivery to pancreatic tumor cells.
Introduction
Pancreatic cancer remains one of the most lethal malignancies in the United States.Citation1,Citation2 The overall median survival after diagnosis is 2–8 months, and the 5-year survival rate is only 3%–6%, indicating the ineffectiveness of current treatment approaches.Citation1–Citation3 It is estimated that nearly 45,220 Americans were diagnosed with pancreatic malignancy, and over 38,460 died from this disease during the past year, marking it as the fourth leading cause of cancer-related death.Citation2,Citation3 This grim scenario has placed a major emphasis on the development of novel therapeutic tools for effective management of this devastating malignancy. In this regard, microRNAs (miRNAs or miRs) have emerged as a novel class of biomolecules that can be exploited clinically, not only as diagnostic and prognostic biomarkers, but also as novel targets for pancreatic cancer therapy.
miRNAs are small noncoding gene-regulatory RNAs that exhibit aberrant expression in several human malignancies, including pancreatic cancer. miRNAs play important roles in cancer initiation, progression, metastasis, and therapeutic resistance through downregulation of pathologically relevant gene targets.Citation4–Citation7 Hence, inhibition of overexpressed oncogenic miRNAs or restitution of downregulated tumor-suppressor miRNAs in cancer cells could provide new therapeutic avenues to treat cancer. Recently, we identified miR-150 as a novel, tumor suppressor miRNA in pancreatic cancer.Citation8 Expression of miR-150 was downregulated in the majority of pancreatic tumor cases, suggesting its restoration could serve as an effective approach for therapy. However, the physicochemical characteristics of miRNAs – namely anionic charge, hydrophilic nature, sensitivity to nuclease degradation in plasma, and inefficient uptake by tumor cells – are major hurdles in achieving their cellular restoration by exogenous means.Citation9,Citation10 Therefore, the application of miRNAs as potential therapeutic agents requires delivery approaches that protect the miRNAs from nucleases and enhance uptake of the miRNAs by the tumor cells.
Two types of gene delivery systems, viral and nonviral, have been developed.Citation11,Citation12 The advantage of using viral vectors is their high transduction efficiency resulting in efficient delivery of the genetic material into the target cells.Citation13 However, the use of viral gene delivery systems is limited by their inherent immunogenicity, low loading capacity, and other quality control issues.Citation11,Citation14,Citation15 On the other hand, nonviral vectors (liposomes, polymer-based systems, and organic or inorganic nanoparticles) are safer for human use, albeit with lower delivery effectiveness.Citation16–Citation19 In this study, we investigated poly (D,L-lactide-co-glycolide) (PLGA)-based nanoformulation for the delivery of miR-150 to pancreatic cancer cells in vitro. PLGA is a biodegradable and biocompatible polymer and has already been approved by United States Food and Drug Administration for human use.Citation20,Citation21 Our studies show that miR-150-loaded nanoformulation (miR-150-NF) resulted in efficient intracellular delivery of miR-150, potent inhibition of its target gene (MUC4), and associated changes in signaling pathways. Lastly, miR-150-NF treatment led to inhibition of pancreatic tumor cell growth and clonogenicity, and suppressed their malignant behavioral properties. Together, our study characterizes a PLGA-based nanovector platform that could potentially be useful for efficient delivery of miR-150 to the pancreatic tumor cells and, thus, pave the way for novel and effective miRNA-based cancer therapeutics.
Materials and methods
Cell culture
The human pancreatic cancer cell lines, Colo-357 and HPAF, were maintained in a humidified atmosphere of 5% CO2 at 37°C as monolayer cultures in Roswell Park Memorial Institute (RPMI) 1640 media (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Lawrence Ville, GA, USA), penicillin (100 units/mL), and streptomycin (100 μg/mL) (Thermo Fisher Scientific). Validation of cell lines was done by short tandem repeats genotyping and/or expression of defined molecular markers (MUC1, MUC4, vimentin, and DPC4). Moreover, cells were continuously monitored for their typical morphology and intermittently tested for mycoplasma using MycoSensor PCR assay kit (Stratagene, La Jolla, CA, USA) as per manufacturer’s protocol.
Preparation of miR-150-loaded PLGA/Polyethylenimine nanoparticles
Ester terminated PLGA (molecular weight: 40 kDa) and PEI (molecular weight: 25 kDa) were purchased from LACTEL Absorbable Polymers (Birmingham, AL, USA). The ratio of lactic acid to glycolic acid in PLGA is 50:50. PLGA (32 mg) and PEI (100 μg) were dissolved in 1 mL of chloroform. A mixture of acetylated bovine serum albumin (4 mg) and miR-150 (283.5 μg) in 0.2 mL of 1X-Tris-ethylenediaminetetraacetic acid (EDTA) buffer was added into organic phase (the molar ratio of PEI and miRNA was ~1:8), vortexed briefly, and then sonicated using a probe sonicator (~3 W, Sonicator XL; Misonix, Inc., Farmingdale, NY, USA) for 30 seconds over an ice bath to form water-in-oil emulsion. The water-in-oil emulsion was then added into 6 mL of 2.5% w/v polyvinyl alcohol solution prepared with 1X Tris-EDTA buffer and sonicated at 9 W for 3 minutes over ice bath. The emulsion was further stirred for 18 hours at ambient conditions, and then for 2 hours under vacuum to remove chloroform. Nanoparticles were recovered by ultracentrifugation (35,000 rpm for 35 minutes at 4°C, Optima TM, LE-80 K; Beckman Coulter Inc., Brea, CA, USA), washed three times (first wash using 1X Tris-EDTA buffer and the next two washes with sterile deionized water), and then lyophilized (FreeZone 4.5; Labconco, Kansas City, MO, USA).
Encapsulation efficiency
The amount of miR-150-loaded in nanoparticles was determined as described previously.Citation22 In brief, the amount of un-entrapped miR-150 mimics in the nanoparticle wash solutions obtained during the formulation step was quantified using the Quant-iT™ PicoGreen Assay kit (Thermo Fisher Scientific, Waltham, MA, USA). Fluorescence resulting from the binding of PicoGreen reagent to miR-150 was measured in a black-bottomed 96-well plate using FLx-800 microplate reader (BIO-TEK Instruments, Winooski, VT, USA). miR-150 loading in nanoparticles was determined by subtracting the total amount of miR-150 recovered in the wash solutions (miR-150w) from the initial amount of miR-150 (miR-150i) added. Encapsulation efficiency was calculated as:
Physicochemical characterization
The surface morphology and size of nanoparticles were characterized by transmission electron microscopy using Technai™ GCitation2 Spirit Bio TWIN (FEI Company, Hillsboro, OR, USA). Hydrodynamic diameter and zeta potential of nanoparticles were measured in distilled water using Delsa™ Nano C (Beckman Coulter Inc.). Zeta potential values were calculated from measured velocities using Smoluchowski equation. Mean hydrodynamic diameters were calculated for size distribution by weight with the assumption of a lognormal distribution.
In vitro release assay
About 1 mg of miR-150-loaded nanoparticles were incubated with 1 mL of release medium (Tris–EDTA buffer with or without 10% FBS, pH 7.4) in a rotary shaker at 100 rpm at 37°C. At predetermined intervals (after every 24 hours for 14 days), the nanoparticle suspension was centrifuged at 7,500 rpm at 4°C for 10 min. Supernatant (1 μL) was analyzed using the nucleic acid measurement application of NanoDrop ND-1,000 spectrophotometer (Thermo Fisher Scientific), and the amount of miRNA was reported as μg/mL. Release medium was used as a control. Samples were taken in triplicate for each time point.
Measurement of delivery efficacy
Pancreatic cancer cells were seeded in six-well plates and grown until they reached 60%–70% confluence. Cells were treated with miR-150 mimics (Ambion, Austin, TX, USA) (with or without Lipofectamine [Thermo Fisher Scientific]), miR-150-NF (0.05 mg, 0.1 mg, 0.2 mg), or blank (unloaded, 0.2 mg) PLGA nanoparticles. After 16 hours of transfection/treatment, media was replaced with complete media and cells were further cultured for 48 hours. Expression level of mature miR-150 was examined using quantitative reverse-transcription polymerase chain reaction (PCR) assay as described in the following section.
RNA isolation and quantitative reverse-transcription PCR assay
TRIzol reagent (Thermo Fisher Scientific) was used to extract total RNA. Complementary DNA was synthesized using the High Capacity complementary DNA Reverse Transcription Kit (Thermo Fisher Scientific) following manufacturer’s instruction. PCR was performed in 96-well plates using SYBRGreen Master Mix (Thermo Fisher Scientific) on an iCycler system (Bio-Rad Laboratories Inc., Hercules, CA, USA). U6 small nuclear RNA was used as an internal control. Relative amounts of miRNA were normalized against U6 small nuclear RNA, and fold change for miRNA was calculated by the 2−ΔΔCt method. All the primers used in this study are described earlier.Citation8 The thermal conditions used for the real-time PCR were as follows: cycle 1, 95°C for 10 minutes; cycle 2 (×40), 95°C for 10 seconds and 58°C for 45 seconds.
Western blot analysis
Cells were processed for protein extraction and western blotting as described earlier.Citation23 Immunodetection was carried out using specific antibodies against MUC4 (8G7), pY1248-HER2 (mouse monoclonal) (Santa Cruz Biotechnology Inc., Dallas, TX, USA), pHER2, pERK1/2 (mouse monoclonal), ERK1/2, FAK and pFAK (rabbit monoclonal) (Epitomics, Burlingame, CA, USA), and β-actin (mouse monoclonal) (Sigma-Aldrich Co., St Louis, MO, USA). All secondary antibodies (Santa Cruz Biotechnology Inc.) were used at 1:2,000 dilutions. Proteins were visualized with the Super Signal West Femto Maximum sensitivity substrate kit (Thermo Fisher Scientific) and the signal was detected using an LAS-3,000 image analyzer (Fuji Photo Film Co., Tokyo, Japan).
Cell growth and clonogenicity assays
For cell growth assay, cells (2×103 cells per well) were treated with miR-150-NF or blank PLGA nanoparticles (0.05 mg each), as described above, and cultured for 5 days. Cell growth was then monitored using WST-1 assay kit (Hoffman-La Roche Ltd., Basel, Switzerland) as per manufacturer’s instructions. The absorbance was measured at a wavelength of 450 nm using a Bio-Rad Benchmark microplate reader (Bio-Rad Laboratories Inc.). For clonogenicity assay, cells were cultured in six-well plates until they reached 60%–70% confluence, and were subsequently treated with miR-150-NF or blank PLGA nanoparticles (0.05 mg each). Following 48 hours transfection, cells were trypsinized and plated in six-well plates at a density of 1×103 cells per well in a regular media for colony formation. After 2 weeks, colonies were fixed with methanol, stained with crystal violet, photographed, and counted using image analysis software (Gene Tools; Syngene, Frederick, MD, USA).
Motility and invasion assays
Cells were treated with miR-150-NF or blank PLGA nanoparticles (0.05 mg each) for 48 hours, trypsinized, and subjected to motility and invasion assays. For motility assay, cells (1×106) were plated in the top chamber of mono-coated polyethylene terephthalate membrane (six-well insert, pore size 8 μm; BD Biosciences, San Jose, CA, USA). For the invasion assay, 2.5×105 cells were plated in the top chamber of the transwell with a Matrigel-coated polycarbonate membrane (24 wells, 0.8 μm; BD Biosciences). Respective media with 10% FBS was added to the lower chamber as a chemoattractant. After 16 hours of incubation, cells remaining on the upper surface of the insert membrane were removed by cotton swab. Cells that had migrated or invaded through the membrane/Matrigel to the bottom of the insert were fixed, stained with Diff-Quick cell staining kit (Siemens Healthcare Diagnostics Inc., Newark, DE, USA), mounted on a slide, photographed using inverted phase contrast microscope, and counted in ten random view fields.
Statistical analysis
Each experiment was performed independently at least three times. All the values were expressed as mean ± standard deviation. Wherever appropriate, the data were also subjected to unpaired two-tailed Student’s t-test. A value of P<0.05 was considered as significant.
Results
Characterization of miR-150-NF
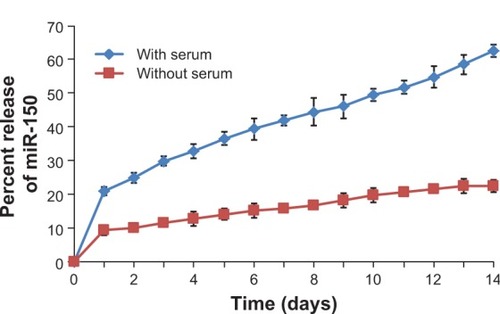
Nanoparticles were characterized for physicochemical properties, namely morphology, size, zeta potential, loading efficiency, and in vitro release profile. Transmission electron microscopy studies revealed that nanoparticles were spherical in shape with a smooth surface and possessed an average particle size of ~168 nm (for blank nanoparticles, ) and ~183 nm (for miR-150-NF, ). Dynamic light scattering studies demonstrated an average hydrodynamic diameter of ~307 nm and a polydispersity of 0.229 for blank nanoparticles, whereas miR-150-NF exhibited average diameter of 399 nm and a polydispersity of 0.177 (data not shown). Nanoparticles had a net negative surface charge and exhibited a zeta potential of −20.22 mV for blank nanoparticles and -20.63 mV for miR-150-NF. Furthermore, our data demonstrate efficient encapsulation of miR-150 mimics (miRNA loading efficiency =78.3%) in PLGA nanoparticles. Additionally, in vitro release profile of miR-150-NF indicated an initial burst release of 9.4% and 21% of the encapsulated miR-150 in serum deprived TE-buffer and serum (10% FBS) containing TE-buffer, respectively, in the first 24 hours followed by a more sustained release over the next 14 days (). These studies also show that miRNA release is faster in the presence of serum (>62% release vs <30% in the absence of serum).
Figure 1 Transmission electron micrographs of PLGA nanoparticles.
Notes: The transmission electron microscopic images of (A) unloaded and (B) miR-150-loaded nanoparticles. Scale bar is 1 μm.
Abbreviation: PLGA, poly (D,L-lactide-co-glycolide).
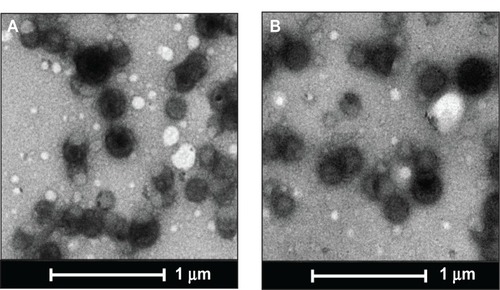
Figure 2 In vitro release profile of miR-150 mimic from miR-150-NF.
Notes: One milligram of miR-150-NF was incubated with 1 mL of release medium (Tris–EDTA buffer with or without 10% FBS, pH 7.4) in a rotary shaker at 100 rpm at 37°C. At 24 hour intervals, the nanoparticle suspension was centrifuged at 7,500 rpm for 10 minutes at 4°C. The amount of miR-150 in the supernatant was determined using ultraviolet spectrophotometry. The experiment was repeated three times and standard deviation was calculated.
Abbreviations: EDTA, ethylenediaminetetraacetic acid; NF, nanoformulation.
miR-150-NF efficiently delivers miR-150 mimics to pancreatic cancer cells
An effective delivery system should possess high transfection efficiency in order to deliver oligonucleotides proficiently into the cells. We evaluated whether the miR-150-NF is able to increase the level of miR-150 in pancreatic cancer cells. Our data revealed that miR-150 expression was significantly increased (~28-fold in Colo-357 and ~26-fold in HPAF cells) in 0.1 mg miR-150-NF (containing 0.69 μg miR-150)-treated cells as compared to those transfected with miR-150–Lipofectamine (containing 0.708 μg miR-150) (). Moreover, miR-150 expression was further increased in a dose-dependent manner (0.05 mg to 0.2 mg) in both the cell lines after treatment with miR-150-NF. Our data suggest that miR-150-NF is highly efficient in delivering miR-150 to pancreatic cancer cells.
Figure 3 miR-150-NF delivers miR-150 mimics to pancreatic cancer cells efficiently.
Notes: Pancreatic cancer cells were seeded in six-well plates to reach 60%–70% confluence. Cells were treated with 0.1 mg of miR-150 mimics (with or without Lipofectamine [Thermo Fisher Scientific, Waltham, MA, USA]), miR-150-NF (0.05 mg [0.245 μg miR-150], 0.1 mg [0.69 μg miR-150], and 0.2 mg [1.38 μg miR-150]), blank (unloaded, 0.1 mg) PLGA nanoparticles. After 16 hours of transfection, medium was replaced with complete media and cells were further cultured for 48 hours. Expression of mature miR-150 was examined in cells by quantitative reverse-transcription-polymerase chain reaction. Comparable expression of miR-150 was observed after treatment of cells with 0.05 mg of miR-loaded NPs and miR-150 mimic (0.708 μg miR-150) with Lipofectamine, whereas significantly higher expression was observed in cells after treatment with miR-150-NF at concentrations of 0.1 mg and 0.2 mg. Results are presented as fold increase in miR-150 expression in various treatments in comparison to miRNA-150 alone. Bars represent mean ± standard deviation, n=3; *P<0.05; **P<0.001; ***P<0.0001.
Abbreviations: miR, microRNA; NF, nanoformulation; NPs, nanoparticles.
![Figure 3 miR-150-NF delivers miR-150 mimics to pancreatic cancer cells efficiently.Notes: Pancreatic cancer cells were seeded in six-well plates to reach 60%–70% confluence. Cells were treated with 0.1 mg of miR-150 mimics (with or without Lipofectamine [Thermo Fisher Scientific, Waltham, MA, USA]), miR-150-NF (0.05 mg [0.245 μg miR-150], 0.1 mg [0.69 μg miR-150], and 0.2 mg [1.38 μg miR-150]), blank (unloaded, 0.1 mg) PLGA nanoparticles. After 16 hours of transfection, medium was replaced with complete media and cells were further cultured for 48 hours. Expression of mature miR-150 was examined in cells by quantitative reverse-transcription-polymerase chain reaction. Comparable expression of miR-150 was observed after treatment of cells with 0.05 mg of miR-loaded NPs and miR-150 mimic (0.708 μg miR-150) with Lipofectamine, whereas significantly higher expression was observed in cells after treatment with miR-150-NF at concentrations of 0.1 mg and 0.2 mg. Results are presented as fold increase in miR-150 expression in various treatments in comparison to miRNA-150 alone. Bars represent mean ± standard deviation, n=3; *P<0.05; **P<0.001; ***P<0.0001.Abbreviations: miR, microRNA; NF, nanoformulation; NPs, nanoparticles.](/cms/asset/4949f46a-5ca6-4ded-a755-c09f77556e46/dijn_a_61949_f0003_c.jpg)
Treatment of miR-150-NF represses the expression of MUC4 and its interacting partner, HER2, and impacts downstream signaling
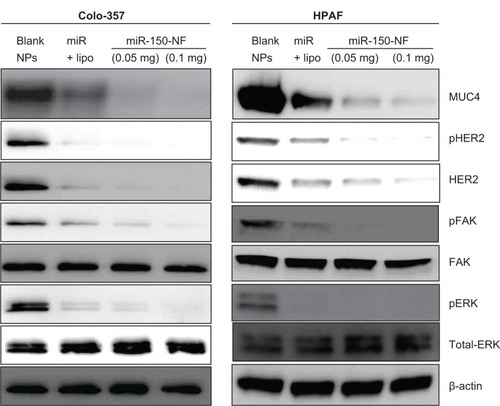
We have shown earlier that MUC4 is a direct target of miR-150.Citation8 Therefore, to delineate the functional efficacy of miR-150-NF, we examined its effect on the expression of MUC4. For this, we treated Colo-357 and HPAF cells with blank nanoparticles (0.1 mg), miR-150-NF (0.05 mg or 0.1 mg), or miR-150 mimics with Lipofectamine for 48 hours and examined the expression of MUC4. Our data reveal that both miR-150-NF and nonformulated miR-150 are able to repress MUC4 expression in Colo-357 and HPAF cells. Interestingly, the data demonstrate that the effect of MUC4 inhibition is more potent when cells are treated with miR-150-NF as compared to treatment with Lipofectamine–miR-150 complex (). We next examined the effect of miR-150-NF treatment on the expression of HER2 as well as its downstream signaling. HER2 has previously been shown by us to physically interact with MUC4, where the latter causes its stabilization.Citation24 The data from immunoblot analysis reveal that the expression of HER2 and its phosphorylated form (pY1248-HER2) is reduced following treatment with miR-150-NF in both the cell lines (). Furthermore, we observed a decreased phosphorylation of ERK and FAK (downstream effectors of HER2) in miR-150-NF-treated cells as compared to blank nanoparticles. Altogether, our data suggest that miR-150-NF represses MUC4 and HER2, leading to attenuation of its downstream signaling in pancreatic cancer cells, thus confirming the functional efficacy of miR-150-NF.
Figure 4 miR-150-NF represses MUC4 expression, HER2 expression, and HER2 downstream signaling in pancreatic cancer cells.
Notes: Colo-357 and HPAF cells were treated with blank NPs (0.1 mg), miR-150-NF (0.05 or 0.1 mg), or miR-150 with Lipofectamine (Thermo Fisher Scientific, Waltham, MA) for 48 hours. Immunoblotting was performed for MUC4, HER2, p-HER2, ERK1/2, pERK1/2, FAK, and pFAK. β-actin was used as a loading control.
Abbreviations: lipo, Lipofectamine; NF, nanoformulation; NPs, nanoparticles; MUC4, Mucin 4; HER2, human epidermal growth factor receptor 2; pHER2, phosphorylated HER2; ERK, extracellular signal-regulated kinase; pERK, phosphorylated ERK; FAK, focal adhesion kinase; pFAK, phosphorylated FAK.
miR-150-NF treatment inhibits growth and clonogenicity of pancreatic cancer cells
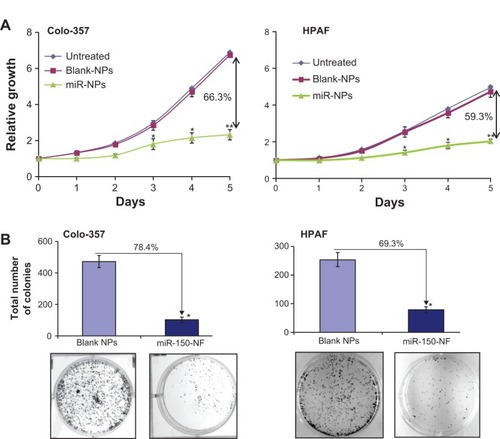
In order to examine the effects of miR-150-NF on pancreatic cancer cell phenotype, we performed a series of in vitro assays. First, we studied the effect of miR-150-NF on the cellular morphology and growth of pancreatic cancer cells. Cells treated with miR-150-NF (0.05 mg and 0.1 mg) showed alterations in morphology as compared to untreated cells (). At 0.1 mg concentration of miR-150-NF, remarkable changes in the morphology of pancreatic cancer cells (Colo-357 as well as HPAF) were observed as illustrated by rounded shape, smaller size, reduced spreading over the culture dish, and detachment from the substratum as compared to the control cells (). Furthermore, the growth kinetics study demonstrated significant growth inhibition in miR-150-NF-treated pancreatic cancer cells (~66% in Colo-357 and ~59% in HPAF) on day 5 as compared to their respective controls (). We next examined the effect of miR-150-NF on the anchorage-dependent clonogenic ability of the pancreatic cancer cells. The data reveal that miR-150-NF decreases the clonogenic ability of Colo-357 and HPAF cells by 78.4% and 69.3%, respectively, as compared to that observed in blank nanoparticle-treated cells (). Thus, our findings suggest a significant antitumor efficacy of miR-150-NF against pancreatic cancer cells.
Figure 5 Treatment of pancreatic cancer cells with miR-150-NF alters the morphology of pancreatic cancer cells.
Notes: Pancreatic cancer cells were seeded in six-well plates and allowed to attain 60%–70% confluence prior to miR-150-NF (0.05 or 0.1 mg/mL) treatment for 48 hours. Representative micrographs are from one of the random fields of view (magnification 100×) of cells.
Abbreviation: NF, nanoformulation.
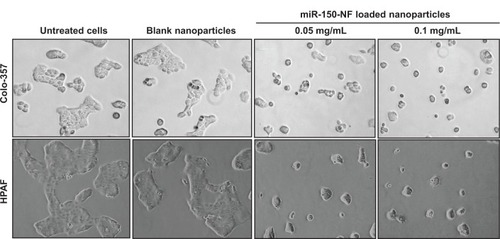
Figure 6 miR-150-NF decreases growth and clonogenicity of pancreatic cancer cells.
Notes: (A) Colo-357 and HPAF cells (2×103 per well) were seeded in 96-well plates. After 24 hours (considered as day 0), cells were treated with either miR-150-NF or blank NPs (0.05 mg each) and growth was monitored by WST-1 assay every day for next 5 days. After analysis, data were presented as relative fold-growth induction compared with growth of cells at day 0. Growth inhibition of cells treated with miR-150-NF was compared to cells treated with blank NPs on day 5 and is shown as percentage. Bars represent mean ± SD (n=3); *P<0.05; **P<0.001. (B) Pancreatic cancer cells were treated with miR-150 mimics-loaded or blank NPs and, 48 hours later, cells were trypsinized and seeded in six-well plates (1×103 cells per well) for clonogenicity assay. After 2 weeks, colonies were stained with 0.1% crystal violet, photographed, and counted using imaging system. Data are presented as percent inhibition of clonogenic ability of miR-150-NF-treated cells as compared with their respective controls. Bars represent the mean of total number of colonies ± SD (n=3); *P<0.05.
Abbreviations: miR, microRNA; NF, nanoformulation; NPs, nanoparticles; SD, standard deviation.
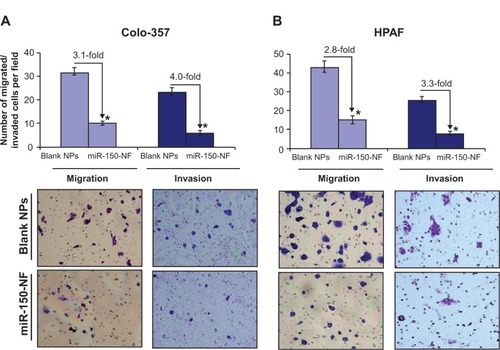
Treatment with miR-150-NF results in the suppression of malignant behavior of pancreatic cancer cells
In our previous studies, we have shown an important role of MUC4 in potentiating the aggressive malignant behavior of pancreatic tumor cells.Citation8 Therefore, we investigated the effect of miR-150-NF treatment on motility and invasive potential of Colo-357 and HPAF cells. Cell motility assay was performed by following the migration of tumor cells under chemotactic drive (FBS) using a Boyden’s chamber. Data show a significant decrease (~3.1- and 2.8-fold in Colo-357 and HPAF cells, respectively) in the number of migrated cells following miR-150-NF treatment as compared to their respective controls (). Furthermore, Matrigel invasion assay was performed to determine the effect of miR-150-NF treatment on invasiveness of pancreatic cancer cells. The data demonstrate that, following treatment with miR-150-NF, invasiveness of the Colo-357 and HPAF cells was decreased ~4.0- and 3.3-fold, respectively, as compared to the cells treated with blank nanoparticles (). Altogether, the data suggest a potent effect of miR-150-NF treatment on attenuating the malignant potential of pancreatic cancer cells.
Figure 7 Treatment with miR-150-NF decreases the motility and invasion of pancreatic cancer cells.
Notes: (A) Colo-357 and (B) HPAF cells were treated with miR-150-NF or blank NPs for 48 hours as described above. Cells were trypsinized and seeded (1×106 and 2.5×105 for migration and invasion, respectively) in serum-deprived media on non-coated or Matrigel-coated membranes for motility and invasion assays, respectively. Medium containing 10% FBS was added in the lower chamber as a chemoattractant and incubated for 16 hours in transwell plates. Cells that had migrated/invaded through the membrane/Matrigel to the bottom of the insert were fixed, stained, photographed using inverted phase contrast microscope, and counted in ten random view fields. Bars represent the mean ± standard deviation (n=3) of number of migrated/invaded cells per field; *P<0.05.
Abbreviations: FBS, fetal bovine serum; NF, nanoformulation; NPs, nanoparticles.
Discussion
miRNA delivery approach, also known as miRNA replacement therapy, is focused on reinstating functional miRNAs that are lost in the diseased cells. In the present study, we generated PLGA-based miR-150-NF to facilitate the delivery of miR-150 into pancreatic cancer cells exhibiting its downregulated expression. Our studies show that miR-150-NF was able to efficiently deliver miR-150 mimics to pancreatic cancer cells. Treatment with miR-150-NF led to the repression of miR-150 gene target, MUC4, and also caused downregulation of its interacting partner, HER2. We also observed suppression of HER2 and its downstream signaling upon treatment with miR-150-NF, which resulted in significant inhibition of pancreatic cancer cell growth, clonogenicity, motility, and invasion.
Nucleotide delivery requires carriers that are safe, are capable of efficient endocytosis into tumor cells, and possibly possess targeting capabilities for delivery to specific cells.Citation25–Citation28 In the present study, we chose the polymer PLGA for the preparation of nanoparticles to deliver miR-150 due to advantages of PLGA over other polymers. PLGA-based polymer matrices exhibit tremendous versatility that allows for their tailoring to meet the specific need.Citation20,Citation29 Furthermore, PLGA polymers are biodegradable and are approved for human use.Citation20,Citation21 In addition, PLGA-based nanoparticles permit sustained intracellular release of encapsulated agents, leading to potent and prolonged therapeutic efficacy.Citation30–Citation32 However, one of the limitations of the PLGA nanoparticles is their low encapsulation efficiency (<50%) for low molecular weight and hydrophilic molecules.Citation22,Citation29 We were able to overcome this limitation by incorporating PEI, a cationic polymer, in the PLGA matrix. PEI has a high positive charge density and, therefore, helps in enhanced encapsulation of negatively charged moieties such as nucleic acids.Citation33,Citation34 Indeed, addition of cationic polymers, such as PEI or chitosan, has been shown previously to improve encapsulation efficiency of small interfering RNAs.Citation35,Citation36
Additional criteria that affect the efficacy of nanoparticles are their size and zeta potential, which determine the level of cellular and tissue uptake.Citation37–Citation39 Various studies have been conducted to investigate the effect of nanoparticle size on gene delivery efficiency. Smaller size nanoparticles exhibit greater gene delivery efficiency, and some studies suggest 400 nm as the effective upper size limit for passive targeting.Citation37,Citation40–Citation42 Although the average hydrodynamic diameter of our miR-150-loaded nanoparticles was close to this upper limit, they still exhibited efficient intracellular delivery of miRNAs. However, it will be desirable to further refine our formulation methodology to reduce the size of miR-150-NF prior to initiating in vivo studies.
Zeta potential is an indicator of the surface charge of nanoparticles and is a key parameter that determines nanoparticle stability in suspension.Citation37,Citation43 Nanoparticles with high negative or positive zeta potential values are generally more stable in dispersion. Despite the addition of PEI, the net zeta potential of miR-150-NF was −20.63 mV. This could be due to the fact that the amount of PEI in the formulation was very small (100 μg) compared to the amount of PLGA (32 mg). Moreover, the presence of the negatively charged miRNA molecules also would have neutralized some of the cationic charge arising from PEI. In additional studies, we observed a slow and sustained release of miR-150 in vitro, which is highly desirable for sustained therapeutic activity.Citation37,Citation44 Faster miRNA release in the presence of serum (>62% release versus <30% in the absence of serum) might be attributed to a higher percentage of polyanions, mainly due to albumins in serum, that can compete with anionic miRNAs in binding to cationic PEI.
miRNAs play important roles in a wide range of biological processes, such as cell proliferation, apoptosis, differentiation, metabolism, and migration and invasion, through regulation of target gene expression.Citation5,Citation45 To be therapeutically effective, miR-150-NF treatment should be capable of inducing both biological and molecular effects. In that regard, we observed that treatment of pancreatic cancer cells with miR-150-NF caused downregulation of miR-150 target gene (MUC4) expression. Additionally, miR-150-NF also repressed the expression of HER2, an interacting partner of MUC4, leading to attenuation of its downstream signaling (FAK and ERK) in pancreatic cancer cells. These observations correlated with decreased growth and clonogenicity of pancreatic tumor cells and suppression of malignant behavioral properties. These effects were consistent with our previous findingsCitation8 and clearly demonstrated the therapeutic efficacy of miR-150-NF in vitro.
In summary, we have successfully developed PLGA-based nanodelivery system of miR-150 that efficiently encapsulates miR-150, demonstrates sustained release, effectively delivers miR-150, and inhibits growth and malignant potential of pancreatic cancer cells. Since PLGA nanoparticles can be easily surface modified,Citation20,Citation25 our formulation also holds immense potential to ensure the targeted delivery of miRNAs to the cancer cells under in vivo setting. Additional studies are, however, needed to further test its effectiveness in vivo. Nonetheless, our data provides proof of the concept for miRNA delivery using PLGA-based nanoformulation.
Acknowledgments
Grant Support: NIH/NCI [CA137513, CA167137, CA175772, CA185490 (to APS) and CA169829, CA186233 (to SS)] and USAMCI.
Disclosure
The authors report no conflicts of interest in this work.
References
- LongJZhangYYuXOvercoming drug resistance in pancreatic cancerExpert Opin Ther Targets20111581782821391891
- SiegelRNaishadhamDJemalACancer statistics, 2013CA Cancer J Clin201363113023335087
- AroraSBhardwajASinghSAn undesired effect of chemotherapy: gemcitabine promotes pancreatic cancer cell invasiveness through reactive oxygen species-dependent, nuclear factor κB- and hypoxia-inducible factor 1α-mediated up-regulation of CXCR4J Biol Chem201319288211972120723740244
- Alvarez-GarciaIMiskaEAMicroRNA functions in animal development and human diseaseDevelopment20051324653466216224045
- BhardwajASinghSSinghAPMicroRNA-based Cancer Therapeutics: Big Hope from Small RNAsMol Cell Pharmacol2010221321921289871
- BushatiNCohenSMmicroRNA functionsAnnu Rev Cell Dev Biol20072317520517506695
- SrivastavaSKBhardwajALeavesleySJGrizzleWESinghSSinghAPMicroRNAs as potential clinical biomarkers: emerging approaches for their detectionBiotech Histochem20138837338723293934
- SrivastavaSKBhardwajASinghSMicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cellsCarcinogenesis2011321832183921983127
- KobayashiEHornicekFJDuanZMicroRNA Involvement in OsteosarcomaSarcoma2012201235973922550419
- PecotCVCalinGAColemanRLLopez-BeresteinGSoodAKRNA interference in the clinic: challenges and future directionsNat Rev Cancer201111596721160526
- IbraheemDElaissariAFessiHGene therapy and DNA delivery systemsInt J Pharm2014459708324286924
- RaziSSBaradaranBLotfipourFKazemiTMohammadnejadLGene therapy, early promises, subsequent problems, and recent breakthroughsAdv Pharm Bull2013324925524312844
- YoungLSSearlePFOnionDMautnerVViral gene therapy strategies: from basic science to clinical applicationJ Pathol200620829931816362990
- ItakaKKataokaKRecent development of nonviral gene delivery systems with virus-like structures and mechanismsEur J Pharm Biopharm20097147548318955136
- SchagenFHOssevoortMToesREHoebenRCImmune responses against adenoviral vectors and their transgene products: a review of strategies for evasionCrit Rev Oncol Hematol200450517015094159
- AignerANonviral in vivo delivery of therapeutic small interfering RNAsCurr Opin Mol Ther2007934535217694447
- MerdanTKopecekJKisselTProspects for cationic polymers in gene and oligonucleotide therapy against cancerAdv Drug Deliv Rev20025471575812204600
- ZhouJShumKTBurnettJCRossiJJNanoparticle-Based Delivery of RNAi Therapeutics: Progress and ChallengesPharmaceuticals (Basel)201368510723667320
- TyagiNRathoreSSGhoshPCEnhanced killing of human epidermoid carcinoma (KB) cells by treatment with ricin encapsulated into sterically stabilized liposomes in combination with monensinDrug Deliv20111839440421438723
- DanhierFAnsorenaESilvaJMCocoRLe BretonAPréatVPLGA-based nanoparticles: an overview of biomedical applicationsJ Control Release201216150552222353619
- LangerRTirrellDADesigning materials for biology and medicineNature200442848749215057821
- PatilYPanyamJPolymeric nanoparticles for siRNA delivery and gene silencingInt J Pharm200936719520318940242
- SinghSSrivastavaSKBhardwajAOwenLBSinghAPCXCL12-CXCR4 signalling axis confers gemcitabine resistance to pancreatic cancer cells: a novel target for therapyBr J Cancer20101031671167921045835
- ChaturvediPSinghAPChakrabortySMUC4 mucin interacts with and stabilizes the HER2 oncoprotein in human pancreatic cancer cellsCancer Res2008682065207018381409
- JainAKDasMSwarnakarNKJainSEngineered PLGA nanoparticles: an emerging delivery tool in cancer therapeuticsCrit Rev Ther Drug Carrier Syst20112814521395514
- KesharwaniPGajbhiyeVJainNKA review of nanocarriers for the delivery of small interfering RNABiomaterials2012337138715022796160
- PanyamJLabhasetwarVBiodegradable nanoparticles for drug and gene delivery to cells and tissueAdv Drug Deliv Rev20035532934712628320
- WangJLuZWientjesMGAuJLDelivery of siRNA therapeutics: barriers and carriersAAPS J20101249250320544328
- BarichelloJMMorishitaMTakayamaKNagaiTEncapsulation of hydrophilic and lipophilic drugs in PLGA nanoparticles by the nanoprecipitation methodDrug Dev Ind Pharm19992547147610194602
- CohenHLevyRJGaoJSustained delivery and expression of DNA encapsulated in polymeric nanoparticlesGene Ther200071896190511127577
- KhanABenboubetraMSayyedPZSustained polymeric delivery of gene silencing antisense ODNs, siRNA, DNAzymes and ribozymes: in vitro and in vivo studiesJ Drug Target20041239340415545089
- PanyamJZhouWZPrabhaSSahooSKLabhasetwarVRapid endo-lysosomal escape of poly(DL-lactide-co-glycolide) nanoparticles: implications for drug and gene deliveryFASEB J2002161217122612153989
- ReadMLSinghSAhmedZA versatile reducible polycation-based system for efficient delivery of a broad range of nucleic acidsNucleic Acids Res200533e8615914665
- ZhuLMahatoRILipid and polymeric carrier-mediated nucleic acid deliveryExpert Opin Drug Deliv201071209122620836625
- PatilYBSwaminathanSKSadhukhaTMaLPanyamJThe use of nanoparticle-mediated targeted gene silencing and drug delivery to overcome tumor drug resistanceBiomaterials20103135836519800114
- TaharaKSakaiTYamamotoHTakeuchiHKawashimaYEstablishing chitosan coated PLGA nanosphere platform loaded with wide variety of nucleic acid by complexation with cationic compound for gene deliveryInt J Pharm200835421021618178349
- DinarvandRSepehriNManoochehriSRouhaniHAtyabiFPolylactide-co-glycolide nanoparticles for controlled delivery of anticancer agentsInt J Nanomedicine2011687789521720501
- GaumetMVargasAGurnyRDelieFNanoparticles for drug delivery: the need for precision in reporting particle size parametersEur J Pharm Biopharm2008691917826969
- TyagiNGhoshPCFolate receptor mediated targeted delivery of ricin entrapped into sterically stabilized liposomes to human epidermoid carcinoma (KB) cells: effect of monensin intercalated into folate-tagged liposomesEur J Pharm Sci20114334335321621613
- KimJHParkJSYangHNThe use of biodegradable PLGA nanoparticles to mediate SOX9 gene delivery in human mesenchymal stem cells (hMSCs) and induce chondrogenesisBiomaterials20113226827820875683
- PrabhaSZhouWZPanyamJLabhasetwarVSize-dependency of nanoparticle-mediated gene transfection: studies with fractionated nanoparticlesInt J Pharm200224410511512204570
- IshidaOMaruyamaKSasakiKIwatsuruMSize-dependent extravasation and interstitial localization of polyethyleneglycol liposomes in solid tumor-bearing miceInt J Pharm1999190495610528096
- HallJBDobrovolskaiaMAPatriAKMcNeilSECharacterization of nanoparticles for therapeuticsNanomedicine (Lond)2007278980318095846
- UtrejaPJainSTiwaryAKNovel drug delivery systems for sustained and targeted delivery of anti-cancer drugs: current status and future prospectsCurr Drug Deliv2010715216120158482
- IorioMVCroceCMMicroRNAs in cancer: small molecules with a huge impactJ Clin Oncol2009275848585619884536

