Abstract
Neuropathic pain is a common and refractory chronic pain that affects millions of people worldwide. Its underlying mechanisms are still unclear, but they may involve long noncoding RNAs (lncRNAs), which play crucial roles in a variety of biological functions, including nociception. We used microarrays to investigate the possible interactions between lncRNAs and neuropathic pain and identified 22,213 lncRNAs and 19,528 mRNAs in the spinal cord in a mouse model of spared nerve injury (SNI)-induced neuropathic pain. The abundance levels of 183 lncRNAs and 102 mRNAs were significantly modulated by both SNI and administration of minocycline. A quantitative real-time polymerase chain reaction analysis validated expression changes in three lncRNAs (NR_015491, ENSMUST00000174263, and ENSMUST00000146263). Class distribution analysis of differentially expressed lncRNAs revealed intergenic lncRNAs as the largest category. Functional analysis indicated that SNI-induced gene regulations might be involved in the activities of cytokines (IL17A and IL17F) and chemokines (CCL2, CCL5, and CCL7), whereas minocycline might exert a pain-alleviating effect on mice through actin binding, thereby regulating nociception by controlling the cytoskeleton. Thus, lncRNAs might be responsible for SNI-induced neuropathic pain and the attenuation caused by minocycline. Our study could implicate lncRNAs as potential targets for future treatment of neuropathic pain.
Introduction
Neuropathic pain is one of the most common and disabling causes of the chronic pain state, affecting up to 8% of the global population.Citation1 Defined as “pain caused by a lesion or disease of the somatosensory system”,Citation2 neuropathic pain may have a diverse array of clinical causes in both the peripheral and the central nervous system (CNS).Citation1 A substantial body of evidence points to several important aberrant pathogeneses that may contribute to neuropathic pain, such as peripheral and central sensitizationCitation3,Citation4 and pathological activation of microglia.Citation5–Citation7 However, little is known about the genetic basis of the mechanisms behind these changes.
Human protein-coding genes represent only ~1%–2% of the genetic material or 25,000 genes.Citation8 This indicates that most eukaryotic genomes are transcribed, but they generate a large population of noncoding RNAs (ncRNAs).Citation9 Based on transcript size, these ncRNAs can be divided into small noncoding RNAs (<200 bp) and long noncoding RNAs (lncRNAs; ≥200 nt). Unlike the well-characterized microRNAs, the majority of small ncRNAs, lncRNAs are often poorly conserved at the sequence level and are considered to be subject to post-transcriptional processing, intracellular or intercellular transport, association with RNA-binding proteins, and myriad interactions with other molecules.Citation10 Therefore, lncRNAs can serve as highly dynamic, modular, multimodal, and energy efficient factors for epigenetic interference within the nucleus and cytoplasm.Citation11
Recent studies have shown that some lncRNAs are preferentially expressed in the nervous system when compared with other organ systemsCitation10,Citation12,Citation13 and that lncRNAs play important roles in diseases like neuropathic pain. For example, Jiang et al have reported a large number of differentially expressed (DE) lncRNAs in the spinal cord following spinal nerve ligation.Citation14 A novel antisense lncRNA, Kcna2, which is also found in dorsal root ganglia (DRG), is thought to participate in the development of neuropathic pain by specifically silencing expression of Kcna2 (which encodes the potassium voltage-gated channel subfamily A member 2).Citation15 Two functional lncRNAs, uc.48+ and NONRATT021972, regulate diabetic neuropathic pain (DNP) through the P2X3 and P2X7 receptors, respectively, in the DRG.Citation16–Citation18 These findings suggest that lncRNAs have an important effect on neuropathic pain.
One treatment for neuropathic pain is minocycline, a second-generation tetracycline antibiotic. This lipophilic molecule is absorbed rapidly and readily across the blood–brain barrier and may act by several possible mechanisms,Citation19,Citation20 including inhibition of immune cell and microglia activation, inhibition of proinflammatory cytokine release, inhibition of nitric oxide release, and augmentation of anti-inflammatory cytokine release. However, recent studiesCitation19,Citation21 have addressed different opinions about the efficacy and the targets being inhibited. Unlike classic medications, such as gabapentin and pregabalin, that have well-known mechanisms operating generally at the protein level, the mechanism by which minocycline attenuates neuropathic pain is still unclear.Citation19 In the present study, we investigated the genome-wide expression of lncRNAs and mRNAs in the spinal cord following spared nerve injury (SNI)-induced neuropathic pain, in an attempt to clarify the gene mechanisms underlying the pathophysiological aberrations that lead to neuropathic pain. We also used minocycline as a treatment control to filter genes that may be DE during the regulation of neuropathic pain, as well as to explore the mechanism of this drug at the gene level. The findings indicate that lncRNAs may represent crucial mediators and potential therapeutic targets for neuropathic pain.
Materials and methods
Animals
All experimental procedures were approved by the Institutional Animal Care and Use Committee of Guangzhou Medical University and were carried out in accordance with the guidelines of the National Institutes of Health on animal care and the ethical guidelines for investigation of experimental pain in conscious animals.Citation44 Male Balb/c mice aged 8 weeks were purchased from Guangdong Medical Laboratory Animal Center (Guangzhou, China). Mice were housed in separated cages and the rooms were kept at 24±1°C (297±1 K) temperature and 50%–60% humidity, under a 12:12 light–dark cycle and with free access to food and water ad libitum. They were numbered and randomly allocated in 3 groups: Sham, vehicle (V)-SNI (vehicle repeatedly administrated before and after SNI), and minocycline (MC)-SNI (minocycline repeatedly administrated before and after SNI).
Behavioral tests
Mice of 3 groups (6 per group) were tested for basal paw withdrawal mechanical thresholds (PWMTs) before and 7 days after modeling. PWMT was assessed with the up-down method described previously,Citation45,Citation46 using a set of von Frey hairs with logarithmically incremental stiffness from 0.04 to 2.04 g (0.04, 0.07, 0.16, 0.4, 0.60, 1.0, 1.4, 2.04 g). The 0.4 g stimulus, in the middle of the series, was applied first. The observation of a positive response (paw lifting, shaking, or licking) within 5 seconds was then followed by an application of a thinner filament (or a thicker one if the response was negative).
Spared nerve injury (SNI) model
SNI surgeries were performed to mice according to previous works.Citation22,Citation23 In brief, mice were anesthetized with an injection of a mixture of chloral hydrate (0.4 g/kg ip). The skin of the lateral thigh was incised, and the biceps femoris muscle was dissected bluntly to expose the left sciatic nerve and its three terminal branches (the sural, common peroneal, and tibial nerves). The common peroneal and the tibial nerves were tightly ligated with 5-0 silk. Then, the nerve was transected distal to the ligation, and 2–4 mm length of nerve fiber was removed. Great care was taken to avoid any contact with or stretching of the intact sural nerve. The wound was closed in two layers. The Sham group was given all procedures except ligation.
Drug administration
Normal saline and minocycline were given intraperitoneally to mice in V-SNI and MC-SNI groups (Sigma-Aldrich Co., St Louis, MO, USA; 30 mg/kg), respectively, as scheduled: 16 and 1 h before surgery, then twice daily for 7 days after SNI.
RNA extraction
After the behavioral tests, the mice were sacrificed by rapid decapitation under anesthesia. After the exposure of the spinal cord with serial laminectomy, the lumbar (L4–L5) segments of spinal cords were transversely sectioned and hemi-dissected along the midline. Only the dorsal half of the lumbar spinal cord in the ipsilateral (operated) side of Sham, V-SNI and MC-SNI mice was collected, snap-frozen in liquid nitrogen, and stored at −80°C (193 K) until analysis. The tissue samples were washed three times with cold PBS and were scraped into Trizol reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. The quality and the concentration of the RNA samples were monitored at absorbance ratios of A260/A280 and A260/230 using a NanoDrop ND-1000 Spectrometer (NanoDrop Technologies, Wilmington, DE, USA). RNA integrity was assessed using standard denaturing agarose gel electrophoresis.
Microarray and computational analysis
The microarray hybridization and analysis were performed by Kangcheng Bio-tech, Shanghai PR China. Briefly, RNA was purified from total RNA after removal of rRNA (mRNA-ONLY Eukaryotic mRNA Isolation Kit, Epicentre). Then, each sample was amplified and transcribed into fluorescent cRNA along the entire length of the transcripts without 39 bias utilizing a random priming method. The labeled cRNAs were hybridized to the Mouse LncRNA Array v3.0 (8×60 K, Arraystar). About 35,923 lncRNAs and 24,881 coding transcripts which were collected from the most authoritative databases such as RefSeq, UCSC Knowngenes, Ensembl, and many related landmark publications can be detected. The arrays were scanned by the Agilent Scanner G2505B, and the acquired array images were analyzed by Agilent Feature Extraction software (version 11.0.1.1). Quantile normalization and subsequent data processing were performed by using the GeneSpring GX v12.1 software package (Agilent Technologies, Santa Clara, CA, USA). Normalized data were log2-transformed and used for comparisons. LncRNAs and mRNAs, that is, at least 3 out of 9 samples have flags in Present or Marginal (“All Targets Value”) were chosen for further data analysis. DE lncRNAs and mRNAs with statistical significance between the two groups were identified through P-value and fold change filtering.
qRT-PCR analysis
To verify the results of lncRNA microarray, reverse transcription was generated using the SuperScript™ III Reverse Transcriptase (Thermo Fisher Scientific) with a Gene Amp PCR System 9700 (Thermo Fisher Scientific). 1.5 μg RNA, 10 uM primer (), and 2.5 mM each deoxyribonucleotide triphosphates, were incubated for 5 minutes at 65°C and then chilled on ice for 2 minutes. 5× First Strand Buffer, 0.1 M DTT (final concentration), 0.3 μL RNase Inhibitor, and 0.2 μl Superscript III RT were then added. The 20 μL reaction was incubated for 60 minutes at 50°C followed by a final incubation at 70°C for 15 minutes for termination. Quantitative PCR was carried out on a real-time detection instrument ViiA 7 Real-time PCR System (Thermo Fisher Scientific) using 2× PCR master mix (Arraystar) at the following conditions: 10 minutes at 95°C, 40 cycles:10 seconds at 95°C, and 60 seconds at 60°C. All the experiments were replicated three times. The relative expression of genes was calculated based on the 2−DDCt method using the mouse housekeeping GAPDH gene as an endogenous control.Citation47 Expression ratios were subjected to a log2 transform to produce fold change data.
Table 1 The primers of 3 lncRNAs used in this study
Functional annotation enrichment analysis
We used the Database for Annotation, Visualization and Integrated Discovery (DAVID) bioinformatics resources version 6.8Citation48,Citation49 (https://david.ncifcrf.gov) to perform GO (http://www.geneontology.org) analyses for mRNAs. The GO project provides a controlled vocabulary to describe gene and gene product attributes in any organism. The ontology covers three domains: biological process, cellular component, and molecular function. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis is a functional analysis that maps genes to KEGG pathways. We applied a Fisher’s exact P-value cutoff <0.05, correcting for multiple testing with the Benjamin’s false discovery rate. Results of both the analyses were presented in enrichment score (−log10(P-value)) sorted descending. Additionally, we used BinGOCitation26 plugin (v3.0.3) in Cytoscape (v3.4.0) to generate a P-value annotation tree, showing enrichments in related annotations.
Statistical analysis
All results were presented as mean ± standard error. Statistical significance was determined by Student’s t-test for 2 group comparison. Comparisons for 3 groups were conducted with analysis of variance and Turkey’s correction. Both calculations were executed using GraphPad Prism software (version 6.0c). The threshold value used to screen DE lncRNAs and mRNAs was set fold change >2.0. P<0.05 was considered statistically significant.
Results
Minocycline significantly attenuated neuropathic pain induced by SNI
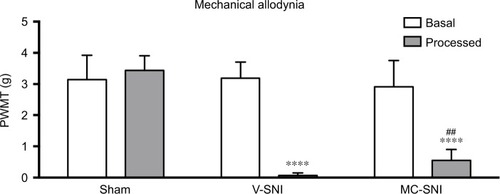
Consistent with previous studies,Citation22,Citation23 the V-SNI group (which received repeated vehicle injections before and after SNI) showed a significant decrease in the PWMTs on the ipsilateral side on day 7, at 0.067±0.035 g, when compared with the preoperative baseline values (3.191±0.219 g, P<0.0001; ) and the values in the sham group (3.438±0.192 g, P<0.0001; ). A significant attenuation in the PWMTs at 7 days after SNI was observed in the MC-SNI group (intraperitoneal [ip] administration of minocycline before and after SNI), at 0.554±0.141 g, when compared to the V-SNI group (P<0.001). No significant difference was noted for the baseline PWMTs in the sham (3.438±0.192 g), V-SNI (3.191±0.211 g), and MC-SNI (2.913±0.344 g) (P>0.05) groups.
Figure 1 The paw withdrawal mechanical thresholds (PWMTs) at baseline and 7 days after modeling are presented as mean ± standard error (n=6). ****P<0.0001 indicates a statistically significant difference when compared to its baseline and Sham group and ##P<0.001 indicates a statistically significant difference when compared to the V-SNI group.
Overview of lncRNA and mRNA expression profiles in the spinal cord after SNI and minocycline treatments
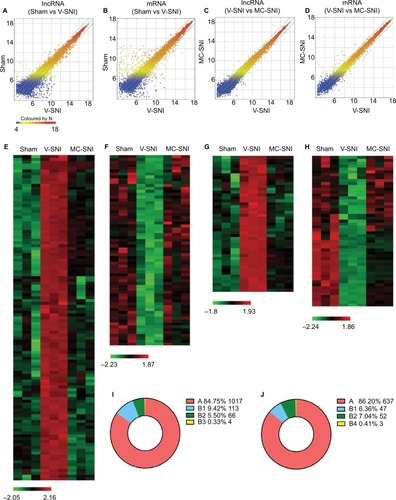
We used microarray technology to obtain an overall profile of lncRNA and mRNA expressions in the ipsilateral L4-L5 spinal cord. The expression features were displayed as scatter plots (). As shown in , a large number of DE lncRNAs and mRNAs were expressed in the V-SNI group compared to the sham group (1,200 DE lncRNAs and 739 DE mRNAs, fold change >2, P<0.05), whereas the difference in the expression of DE lncRNAs and mRNAs was smaller between the V-SNI and MC-SNI groups (). Among these DE transcripts, 183 lncRNAs and 102 mRNAs were greatly altered (fold change >2, P<0.05) in both the sham versus V-SNI and the V-SNI versus MC-SNI groups. These data suggest that SNI led to a substantial alteration in gene expression of lncRNAs and mRNAs, whereas the use of minocycline caused only a slight alteration.
Figure 2 Scatter plots compare the expression of lncRNAs and mRNAs between the sham versus V-SNI group (A and B), and V-SNI versus MC-SNI group (C and D). The values of X and Y axes in the scatter plot are the averaged normalized values in each group (log2 scaled). The green lines indicate a fold change value of 2.0. The transcripts above the top green line and below the bottom green line indicate more than 2-fold changes between pairs. In total, 113 lncRNAs in B1 and 66 in B2 were listed in heat maps E and F; and 47 mRNAs in B1 and 52 in B2 were listed in heat maps G and H. In the heat maps, each column represents a microarray group, while each row represents a regulated gene. The color scale illustrates the relative expression level of lncRNAs: red denotes rich abundance and green denotes poor abundance. Percentages of each pattern for lncRNAs and mRNAs were shown in donut charts (I and J). Pattern A included genes that were modulated only by SNI, whereas pattern B represented genes that were modulated by both SNI and repeated minocycline administration. B1 - genes upregulated by SNI and then downregulated by minocycline, B2 - genes downregulated by SNI and then upregulated by minocycline, B3 - genes upregulated by SNI and further upregulated by minocycline, and B4 - genes downregulated by SNI and further downregulated by minocycline.
The DE transcripts between the sham and V-SNI groups aided in the filtering of lncRNAs and mRNAs that were altered following SNI. The comparison between the V-SNI and MC-SNI groups aided in filtering genes that were DE due to minocycline. The results from these three groups, when taken together, enabled a more accurate identification of genes associated with SNI-induced neuropathic pain.
Gene regulation under two different circumstances was evaluated by dividing the DE genes into two major expression patterns: A and B. Pattern A included genes that were modulated only by SNI, whereas pattern B represented genes that were modulated by both SNI and repeated minocycline administration. Specifically, B1 represented genes that were upregulated by SNI and then downregulated by minocycline; B2 genes were those downregulated by SNI and then upregulated by minocycline; B3 genes were upregulated by SNI and further upregulated by minocycline, and B4 genes were downregulated by SNI and further downregulated by minocycline.
Behavioral tests indicated that minocycline had an alleviating effect on the PWMTs, suggesting that it partially counteracted the effects of SNI. Genes that had an adverse effect on regulation (patterns B1 and B2) could be more responsible for this effect. The gene classification identified 1,017 lncRNAs in pattern A, 113 in B1, 66 in B2, and 4 in B3. The heat maps of DE lncRNAs in B1 and B2 are shown in , respectively.
Among the 739 DE mRNAs detected after SNI, 102 mRNAs were modulated by repeated minocycline administration and were categorized based on the four B-type expression patterns. In total, 47 mRNAs were classified as B1, 52 as B2, and 3 as B4. The levels of the remaining 637 mRNAs were not modulated by minocycline (pattern A). The heat maps of DE mRNAs for the B1 and B2 mRNAs are shown in , respectively. The pattern distribution for lncRNAs and mRNAs are shown in and , respectively.
Quantitative real-time polymerase chain reaction (qRT-PCR) validation of 3 lncRNAs
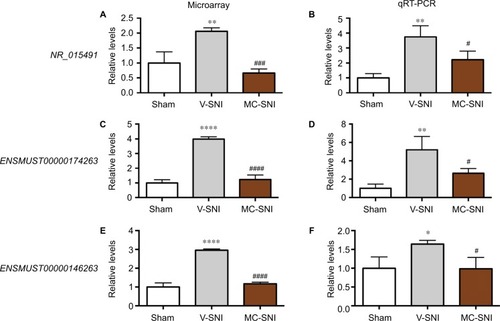
The reliability of the microarray results was validated by qRT-PCR analysis of lncRNAs NR_015491, ENSMUST00000174263, and ENSMUST00000146263.
NR_015491
In the spinal cord, an upregulation of NR_015491 (1.00±0.22 vs 2.06±0.07, P<0.01) was observed 7 days after SNI. Minocycline significantly diminished the level of NR_015491 by 0.67±0.08 (P<0.001, ). The qRT-PCR analyses confirmed the changes detected by the microarray. The quantity of NR_015491 was upregulated by 3.75±0.43 (P<0.01) and downregulated by 2.22±0.33 (P<0.05) in the MC-SNI group ().
Figure 3 The fold changes of the detected lncRNAs NR_015491, ENSMUST00000174263, and ENSMUST00000146263 using microarray (A, C, and E) and qRT-PCR (B, D, and F) were listed. The statistical analysis for each comparison was performed using ANOVA and Turkey’s correction. *P<0.05, **P<0.01, and ****P<0.0001 indicate a statistically significant difference when compared to the sham group; #P<0.05, ###P<0.001, and jpr-10-2503P<0.0001 indicate a significant difference when compared to V-SNI.
ENSMUST00000174263
The microarray results indicated a fold change increase of 3.99±0.09 (P<0.0001) in V-SNI group and a decrease of 1.23±0.18 in the MC-SNI group (P<0.0001; ). The qRT-PCR analyses indicated a fold change increase in ENSMUST00000174263 by 5.21±0.84 (P<0.01) in the V-SNI group and a decrease of 2.66±0.30 (P<0.05) in the MC-SNI group (), in agreement with the microarray data.
ENSMUST00000146263
The microarray analyses indicated a fold change increase in the V-SNI group (from 1.00±0.12 to 2.96±0.04, P<0.0001) and a fold decrease of 1.17±0.04 in the MC-SNI group (P<0.0001; ). The qRT-PCR analysis indicated a fold increase for ENSMUST00000146263 of 1.64±0.06 (P<0.05) in the V-SNI group and a fold decrease of 0.99±0.17 (P<0.05) in the MC-SNI group (), confirming microarray data.
Class distribution analyses of DE lncRNAs
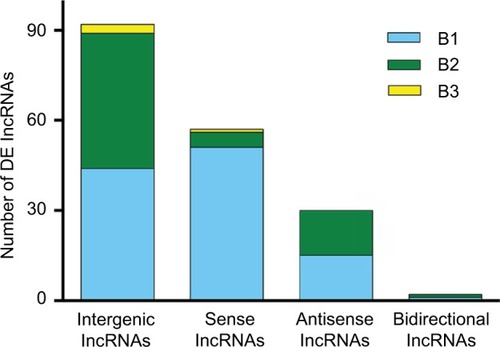
The DE lncRNAs were classified into 4 categories based on their genomic proximity to protein-coding genes:Citation24,Citation25 1) intergenic lncRNAs (lincRNAs), 2) bidirectional lncRNAs, 3) sense lncRNAs, and 4) antisense lncRNAs. The lincRNAs are found in regions that are free of other genes. The bidirectional lncRNAs are oriented head-to-tail to a coding transcript within 1 kb. The sense lncRNAs are transcribed from the sense strand of protein-coding genes and contain exons from protein-coding genes; they may overlap with part of the protein-coding genes or cover the entire sequence of the protein-coding gene. Antisense lncRNAs, by contrast, are transcribed from the antisense strand of protein-coding genes. As shown in , lincRNAs accounted for most of the 183 DE lncRNAs modulated by both SNI and minocycline (pattern B): we identified 92 lincRNAs, 59 sense lncRNAs, 30 antisense lncRNAs, and 2 bidirectional lncRNAs.
Figure 4 Distribution of four lncRNA classes. The distribution of intergenic, sense, antisense, and bidirectional lncRNAs is identified. Blue, green, and yellow colors indicate the number of DE lncRNAs with expression pattern B1, B2, or B3 in each class. Pattern B represented genes that were modulated by both SNI and repeated minocycline administration. B1 - genes upregulated by SNI and then downregulated by minocycline, B2 - genes downregulated by SNI and then upregulated by minocycline, and B3 - genes upregulated by SNI and further upregulated by minocycline.
Functional prediction of DE mRNAs
We explored the possible biological functions that were altered after SNI by focusing on the 739 DE mRNAs identified by comparing the sham and V-SNI groups. The expressions of 390 mRNAs were upregulated (P<0.05, fold change >2.0), and the remaining 349 were downregulated (P<0.05, fold change >2.0). Performance of gene ontology (GO) and the KEGG pathway analyses showed that chemokine activity, chemokine receptor binding, and cytokine activity were among the most enriched GO molecular function annotations of the upregulated mRNAs (; ). A P-value annotation tree was generated using the BinGOCitation26 plugin in Cytoscape with these mRNAs. Part of the tree, shown in , revealed a relationship between the ending annotation nodes, like chemokine activity, as well as the abundant enrichment of their parent annotations (eg, protein binding, receptor binding, and growth factor binding), indicating a concentrative functional change at the gene level.
Figure 5 (A) The enriched molecular function annotations for DE mRNAs from sham versus V-SNI groups. (B) Part of the P-value annotation tree was displayed to reveal chemokine activity and its greatly enriched related annotations. (C and D) The enriched molecular function and biological process annotations for DE mRNAs from V-SNI versus MC-SNI. GO results are presented as descending enrichment scores.
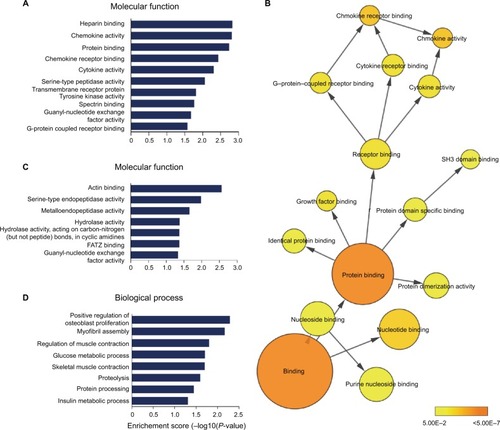
Table 2 Molecular function annotation enrichment results comparing sham versus V-SNI
The transcripts that showed obvious dysregulation by SNI and minocycline (fold change >2 in both the circumstances), which might more strongly reflect a minocycline effect, were also enriched in the GO and KEGG annotations. These transcripts were mainly attributed to the application of minocycline and therefore attracted our interest the most. The paucity of DE mRNAs identified between the V-SNI and MC-SNI groups prompted the inclusion of the genomic proximity-related mRNAs of the DE lncRNAs (V-SNI vs MC-SNI) in the analysis.
Actin binding, serine-type endopeptidase, and hydrolase activities were among the most enriched molecular function annotations (; ). Osteoblast proliferation, myofibril assembly, muscle contraction, and glucose and insulin processes were positively regulated and were the most enriched biological process annotations ().
Table 3 Molecular function annotation enrichment results comparing V-SNI versus MC-SNI
Discussion
Very few studies have focused on neuropathic pain and lncRNAs, despite the strong scientific interest in lncRNAs. To our knowledge, this is the first study to identify global expression changes in lncRNAs in the spinal cord following SNI and minocycline treatment and to explore the possible mechanisms underlying neuropathic pain.
Behavioral tests, performed on all mice before modeling and sampling, confirmed previous findingsCitation22,Citation23 that minocycline alleviates SNI-induced allodynia (). Microarray analysis identified 183 lncRNAs and 102 mRNAs that could represent potential therapeutic targets for minocycline (). The qRT-PCR analysis showed changes in expression levels of NR_015491, ENSMUST00000174263, and ENSMUST00000146263 that were consistent with the microarray results (), thereby validating the microarray data. Similarly, Jiang et al reported 511 DE (fold change >2) lncRNAs (366 upregulated and 145 downregulated) in a spinal nerve ligation-induced neuropathic pain model.Citation14 Therefore, lncRNAs may be involved in neuropathic pain processing.
The vast majority of lncRNA functions are unclear, but emerging evidence now suggests that some lncRNAs play critical roles in neuropathic pain. For example, the lncRNA NONRATT021972, a small interfering RNA, may suppress the upregulation of the P2X3 receptor to reduce the hyperalgesia potentiated by the proinflammatory cytokine TNF-α in type 2 diabetes mellitus rats.Citation17 This lncRNA may also decrease the expression levels of P2X7 mRNA and protein, inhibit the activation of satellite glial cells, and reduce the release of TNF-α, thereby inhibiting the excitability of DRG neurons and decreasing mechanical and thermal hyperalgesia in these rats.Citation18 An antisense lncRNA, Kcna2, is also associated with excitability in DRG neurons through modulation of the Kv current.Citation15 The lncRNA uc.48+, another small interfering RNA, may alleviate DNP by inhibiting the excitatory transmission mediated by the P2X3 receptor in the DRG.Citation16 Further studies are needed to confirm the roles of these DE lncRNAs in neuropathic pain.
Most DE lncRNAs with pattern B were categorized as intergenic (), as they arise from genomic loci that are located far from other annotated genes.Citation27 These lincRNAs are reported to affect the expression of their neighboring genes through different types of mechanisms.Citation27–Citation29 However, in the present case, none of the neighboring or overlapping protein-coding genes were simultaneously altered, indicating that posttranscriptional or translational interference might be more responsible for the regulation.
In our study, we also examined the expression profile of mRNAs by microarrays. Bioinformatics analyses for mRNAs upregulated after SNI showed that a large number of molecular functions of the affected genes were implicated in cytokine and chemokine activities (). Pro-nociceptive factors released from neurons and non-neuronal cells are known to sensitize the neuron of the first pain synapseCitation30 and mediate multistep responses to counteract foreign insults; hence, they play vital roles in the development and maintenance of chronic pain.Citation31,Citation32 Our microarray analysis revealed two cytokine protein-coding transcripts (IL17A and IL17F) among the significantly upregulated mRNAs. Furthermore, a number of CC and CXC chemokine transcripts (eg, CCL2, CCL5, CCL7, CXCL10, and CXCL12) were also identified (). These results supported the classical theory that cytokine and chemokine dysregulations are involved in neuropathic pain through the enhancement of nociceptive transmission.
Minocycline, as an effective pain alleviator, was employed as a treatment control to allow us to filter transcripts that were more related to neuropathic pain. Its use also allowed us to further explore its exact mechanisms. The DE lncRNAs and mRNAs with pattern B were apparently most related to the genomic regulatory functions of this drug. Bioinformatics analysis demonstrated an enrichment of minocycline-induced DE mRNAs with pattern B and that these had molecular function annotations in actin-binding and intracellular enzyme activities (). Thus, minocycline might exert its effects by regulating actin functions at the gene level. Actin binding is believed to regulate spine morphology and synaptic function through the cytoskeleton,Citation33,Citation34 thereby controlling nociception function.Citation35 A previous studyCitation36 addressed the link between impairment of microtubule dynamics and synaptic plasticity in the spinal dorsal horn and the occurrence of paclitaxel-induced painful neuropathy. Pregabalin weakened the formation of web-like filamentous actin (F-actin) and affected cytoskeleton proteins,Citation37 suggesting that chronic pain was associated with changes in the cytoskeleton array and protein folding. The RhoA/LIMK/cofilin pathway is known to modulate the levels of F-actinCitation38 and control the stabilization of actin cytoskeleton. Neurite outgrowth is closely connected to modulations of the cytoskeleton, and microtubule and actin dynamics in particular;Citation39 therefore, actin-binding activity may be pivotal in these regulations. Our microarray analysis detected statistically significant modulation of LIMK1 mRNA, but the fold changes were not >2. Nevertheless, these results supported the possibility of minocycline regulation of actin functions.
Bioinformatics prediction provides a first glance at the functions of the DE mRNAs. However, the limited evidence precluded establishment of the actual role of minocycline, and several limitations should be considered when interpreting our results. First, we used bioinformatics methods solely to predict the possible biological functions that DE mRNAs might participate in. Subsequent studies based on this work might use other methods to validate the possible pathways involved. Second, we sampled spinal dorsal horn tissue only, as this is a site of interest to us in our research. Nevertheless, other areas in the CNS are related to neuropathic pain (eg, the anterior cingulate cortex,Citation40 the ventral tegmental area,Citation41 and the mesencephalic periaqueductal grayCitation42,Citation43). Further studies might include these other areas and compare the identified lncRNAs with the ones from this work. Taken together, the results from the present work have revealed SNI- and minocycline-induced gene alterations, which might provide new verification of the role for lncRNAs as crucial mediators and potential therapeutic targets for the treatment of neuropathic pain.
Conclusion
In summary, microarray technology allowed us to identify and study DE lncRNAs and mRNAs associated with SNI-induced neuropathic pain. SNI-induced gene regulations might be involved in cytokine (IL17A and IL17F) and chemokine (CCL2, CCL5, and CCL7) activities. Minocycline might exert its pain-alleviating effect on mice through actin binding, thereby regulating nociception via control of the cytoskeleton. In the near future, ongoing research is expected to elucidate the roles of individual lncRNAs. Collectively, the results of the present study provide a first step toward future research into effective and safe genetic therapies for the treatment of neuropathic pain.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (number 31100805) and the Project of Science and Technology New Star in Zhujiang Guangzhou City (number 2012J2200036).
Disclosure
The authors report no conflicts of interest in this work.
References
- GilronIBaronRJensenTNeuropathic pain: principles of diagnosis and treatmentMayo Clin Proc201590453254525841257
- JensenTSBaronRHaanpaaMA new definition of neuropathic painPain2011152102204220521764514
- von HehnCABaronRWoolfCJDeconstructing the neuropathic pain phenotype to reveal neural mechanismsNeuron201273463865222365541
- WoolfCJCentral sensitization: implications for the diagnosis and treatment of painPain20111523 SupplS2S1520961685
- WatkinsLRMilliganEDMaierSFSpinal cord glia: new players in painPain200193320120511514078
- WatkinsLRMilliganEDMaierSFGlial activation: a driving force for pathological painTrends Neurosci200124845045511476884
- WatkinsLRMaierSFGlia: a novel drug discovery target for clinical painNat Rev Drug Discov200321297398514654796
- LiangFHoltIPerteaGKaramychevaSSalzbergSLQuackenbushJGene index analysis of the human genome estimates approximately 120,000 genesNat Genet200025223924010835646
- CostaFFNon-coding RNAs: meet thy mastersBioessays201032759960820544733
- QureshiIAMehlerMFLong non-coding RNAs: novel targets for nervous system disease diagnosis and therapyNeurotherapeutics201310463264623817781
- BatistaPJChangHYLong noncoding RNAs: cellular address codes in development and diseaseCell201315261298130723498938
- LipovichLTarcaALCaiJDevelopmental changes in the transcriptome of human cerebral cortex tissue: long noncoding RNA transcriptsCereb Cortex20142461451145923377288
- PonjavicJOliverPLLunterGPontingCPGenomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brainPLoS Genet200958e100061719696892
- JiangBCSunWXHeLNCaoDLZhangZJGaoYJIdentification of lncRNA expression profile in the spinal cord of mice following spinal nerve ligation-induced neuropathic painMol Pain2015114326184882
- ZhaoXTangZZhangHA long noncoding RNA contributes to neuropathic pain by silencing Kcna2 in primary afferent neuronsNat Neurosci20131681024103123792947
- WangSXuHZouLLncRNA uc.48+ is involved in diabetic neuropathic pain mediated by the P2X3 receptor in the dorsal root gangliaPurinergic Signal201612113914826686228
- PengHZouLXieJlncRNA NONRATT021972 siRNA decreases diabetic neuropathic pain mediated by the P2X3 receptor in dorsal root gangliaMol Neurobiol201754151152326742527
- LiuSZouLXieJLncRNA NONRATT021972 siRNA regulates neuropathic pain behaviors in type 2 diabetic rats through the P2X7 receptor in dorsal root gangliaMol Brain201694427107575
- MollerTBardFBhattacharyaACritical data-based re-evaluation of minocycline as a putative specific microglia inhibitorGlia201664101788179427246804
- ZemkeDMajidAThe potential of minocycline for neuroprotection in human neurologic diseaseClin Neuropharmacol200427629329815613934
- SumitaniMUedaHHozumiJMinocycline does not decrease intensity of neuropathic pain intensity, but does improve its affective dimensionJ Pain Palliat Care Pharmacother2016301313525700217
- RichnerMBjerrumOJNykjaerAVaegterCBThe spared nerve injury (SNI) model of induced mechanical allodynia in miceJ Vis Exp201154 pii:3092
- DecosterdIWoolfCJSpared nerve injury: an animal model of persistent peripheral neuropathic painPain200087214915810924808
- MaLBajicVBZhangZOn the classification of long non-coding RNAsRNA Biol2013106924933
- PontingCPOliverPLReikWEvolution and functions of long noncoding RNAsCell2009136462964119239885
- MaereSHeymansKKuiperMBiNGO: a cytoscape plugin to assess overrepresentation of gene ontology categories in biological networksBioinformatics200521163448344915972284
- UlitskyIBartelDPlincRNAs: genomics, evolution, and mechanismsCell20131541264623827673
- RinnJLChangHYGenome regulation by long noncoding RNAsAnnu Rev Biochem20128114516622663078
- MattickJSRinnJLDiscovery and annotation of long noncoding RNAsNat Struct Mol Biol20152215725565026
- ClarkAKOldEAMalcangioMNeuropathic pain and cytokines: current perspectivesJ Pain Res2013680381424294006
- JiRRXuZZGaoYJEmerging targets in neuroinflammation-driven chronic painNat Rev Drug Discov201413753354824948120
- ZhangZJCaoDLZhangXJiRRGaoYJChemokine contribution to neuropathic pain: respective induction of CXCL1 and CXCR2 in spinal cord astrocytes and neuronsPain2013154102185219723831863
- EthellIMPasqualeEBMolecular mechanisms of dendritic spine development and remodelingProg Neurobiol200575316120515882774
- CingolaniLAGodaYActin in action: the interplay between the actin cytoskeleton and synaptic efficacyNat Rev Neurosci20089534435618425089
- WoolfCJSalterMWNeuronal plasticity: increasing the gain in painScience200028854721765176910846153
- ChenSRZhuLChenHWenLLaumetGPanHLIncreased spinal cord Na(+)-K(+)-2Cl(−) cotransporter-1 (NKCC1) activity contributes to impairment of synaptic inhibition in paclitaxel-induced neuropathic painJ Biol Chem201428945311113112025253692
- ParkSLeeJProteomic analysis to identify early molecular targets of pregabalin in C6 glial cellsCell Biol Int2009341273319947953
- MaekawaMIshizakiTBokuSSignaling from Rho to the actin cytoskeleton through protein kinases ROCK and LIM-kinaseScience1999285542989589810436159
- ZulaufLCosteOMarianCMoserCBrenneisCNiederbergerECofilin phosphorylation is involved in nitric oxide/cGMP-mediated nociceptionBiochem Biophys Res Commun200939041408141319896457
- JohansenJPFieldsHLManningBHThe affective component of pain in rodents: direct evidence for a contribution of the anterior cingulate cortexProc Natl Acad Sci U S A200198148077808211416168
- OzakiSNaritaMNaritaMSuppression of the morphine-induced rewarding effect in the rat with neuropathic pain: implication of the reduction in mu-opioid receptor functions in the ventral tegmental areaJ Neurochem20028251192119812358766
- Rodriguez-MunozMSanchez-BlazquezPVicente-SanchezABerrocosoEGarzonJThe mu-opioid receptor and the NMDA receptor associate in PAG neurons: implications in pain controlNeuropsychopharmacology201237233834921814188
- OwenSLGreenALNandiDDBittarRGWangSAzizTZDeep brain stimulation for neuropathic painActa Neurochir Suppl200797Pt 2111116
- ZimmermannMEthical guidelines for investigations of experimental pain in conscious animalsPain19831621091106877845
- ChaplanSRBachFWPogrelJWChungJMYakshTLQuantitative assessment of tactile allodynia in the rat pawJ Neurosci Methods199453155637990513
- DixonWJEfficient analysis of experimental observationsAnnu Rev Pharmacol Toxicol1980204414627387124
- LivakKJSchmittgenTDAnalysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) MethodMethods200125440240811846609
- Huang daWShermanBTLempickiRASystematic and integrative analysis of large gene lists using DAVID bioinformatics resourcesNat Protoc200941445719131956
- Huang daWShermanBTLempickiRABioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene listsNucleic Acids Res200937111319033363
