Abstract
With the introduction of new genetic techniques such as genome-wide array comparative genomic hybridization, studies on the putative genetic etiology of schizophrenia have focused on the detection of copy number variants (CNVs), ie, microdeletions and/or microduplications, that are estimated to be present in up to 3% of patients with schizophrenia. In this study, out of a sample of 100 patients with psychotic disorders, 80 were investigated by array for the presence of CNVs. The assessment of the severity of psychiatric symptoms was performed using standardized instruments and ICD-10 was applied for diagnostic classification. In three patients, a submicroscopic CNV was demonstrated, one with a loss in 1q21.1 and two with a gain in 1p13.3 and 7q11.2, respectively. The association between these or other CNVs and schizophrenia or schizophrenia-like psychoses and their clinical implications still remain equivocal. While the CNV affected genes may enhance the vulnerability for psychiatric disorders via effects on neuronal architecture, these insights have not resulted in major changes in clinical practice as yet. Therefore, genome-wide array analysis should presently be restricted to those patients in whom psychotic symptoms are paired with other signs, particularly dysmorphisms and intellectual impairment.
Introduction
It has become obvious that the phenotypical presentation of psychotic disorders such as schizophrenia is extremely heterogeneous and that their symptoms may comprise the entire spectrum of psychopathology with high interindividual variation.Citation1,Citation2 Although the diagnostic boundaries of schizophrenia still remain unclear, the worldwide lifetime prevalence of this psychotic disorder is estimated to be 0.5%–1%.Citation3 Previous studies have shown a heritability varying from 40%–70%,Citation4 and a family history of schizophrenia has been demonstrated to be the far most important risk factor.Citation5,Citation6 Over the past years, substantial evidence has emerged concerning the relevance of gene–environment interactions for the onset and course of schizophrenia.Citation7,Citation8
With the ongoing developments in genetic techniques, particularly array-based comparative genomic hybridization analysis, it has become possible to investigate the human genome in far greater detail than is possible with routine cytogenetic analysis, leading to the discovery of previously undetectable defects.Citation9 These so-called copy number variants (CNVs) are deletions or duplications ranging in size from 1 kb to 3 Mb resulting in loss or gain of a DNA segment. De novo microdeletions are generally considered of clinical significance and are frequently associated with intellectual disability.Citation10 Of these patients, a substantial percentage develop symptoms from the affective and/or psychotic cluster after adolescence. Microdeletions and duplications may also be causatively involved in patients with normal intelligence and psychiatric disorders like schizophreniaCitation11,Citation12 or autism.Citation13,Citation14 In his review, KirovCitation15 calculated a collective rate of 3% of patients with schizophrenia in whom rare deletions in 1q42, 22q11, and 1q21 and duplications in 16p are found. In the general population, this percentage is estimated to be 0.1%–1%.Citation16 The explanatory power of CNVs for the pathophysiology of psychiatric disorders in general and for their phenotypical presentation, however, still needs to be clarified.Citation17,Citation18
In the present study, genome-wide array analysis was performed in a group of patients with carefully diagnosed psychotic disorders in order to detect possible pathogenic CNVs. If present, their phenotypical relevance was explored and discussed.
Methods
Patients and assessments
Over a period of 30 months, 100 patients referred to the Vincent van Gogh Institute for Psychiatry for treatment of psychotic symptomatology were investigated. Patients with previously demonstrated cytogenetic aberrations and/or intellectual disability (IQ < 70) were excluded. According to the medical ethical guidelines, 80 patients gave written informed consent and 20 refused further participation (CCMO registration number: NL20469.097.07). Assessment of symptomatology and collection of data from history was performed using Comprehensive Assessment of Symptoms and History (CASH),Citation19 Positive and Negative Syndrome Scale (PANSS),Citation20 and Clinical Global Impression Scale (CGI).Citation21 Final clinical diagnoses were made in a so-called Longitudinal Evaluation using All Data (LEAD) conference.Citation22 Subsequently, patients were classified according to the International Statistical Classification of Disease and Related Health Problems, 10th Revision (ICD-10) criteria.Citation23 The main characteristics of the patients including the classification of their psychotic disorders are presented in . Pictures were taken of all patients for evaluation of dysmorphisms by an experienced clinical geneticist (IF). In case CNVs were found by array, the patient was examined in detail by the clinical geneticist.
Table 1 Main characteristics of the patients (n = 80)
Genetic analyses
DNA was isolated from an ethylenediaminetetraacetic acid (EDTA) blood sample and genome wide array analysis was performed with an average resolution of 200 kb using the Affymetrix 250 k SNP array platform (Affymetrix, Santa Clara, CA) as described previously.Citation24
Results
As shows, 50 patients fulfilled the criteria for schizophrenia (F20) and in 11 patients, a diagnosis of acute and transient psychotic disorder was made (F23). A diagnosis of schizoaffective disorder (F25) was established in six patients whereas in another six, bipolar affective disorder, current episode manic with psychotic symptoms, was present (F31.2). In the remaining seven patients, various diagnoses were made (F29, n = 1; F28, n = 2; F22, n = 3; F21, n = 1). All patients received antipsychotic medication according to the hospital standards.
If screening for dysmorphic features suggested a particular monogenetic disorder, specific genetic tests were performed. However, no abnormalities were disclosed. In 77 patients, microarray analysis did not reveal any abnormalities. In three patients, a submicroscopic chromosome imbalance was detected.
Patient A presented with a subacute onset of psychotic symptoms (total scores on PANSS and CGI-S were 79 and 4, respectively) that remitted within 1 month after treatment with 2.5 mg haloperidol. Her history disclosed no major somatic or psychiatric diseases and there was no family load with neuropsychiatric or genetic disorders. In her, an interstitial loss of 3.5 Mb in 1q21.1 comprising 45 genes was found (). Patient B had a history of a mild and short-lasting psychotic episode 1 year before and did not use any psychotropics. Apart from atypical psychotic symptoms, there were mild manic symptoms (total scores on PANSS and CGI-S were 78 and 5, respectively). With a treatment regimen comprising risperidone (4 mg daily) and lithium (1600 mg daily), all symptoms gradually resolved within 3 months. Here, a 375 kb interstitial gain in band p13 of chromosome 1 encompassing two genes was detected (). Patient C had neither a history of any psychiatric or somatic disease nor a family load with neuropsychiatric or genetic disorders. Shortly prior to admission he developed paranoid psychotic symptoms (total scores on PANSS and CGI-S were 63 and 2, respectively) which fully remitted after 1 month of treatment with risperidone 4 mg daily. He had a 1.1 Mb interstitial gain in 7q11.21q11.22 containing 11 genes ().
Figure 1 In the upper panel (i) of each plot (A–C), the log2 test over reference ratio is plotted on the y-axis against the genomic Mb position from pter to qter on the respective chromosome represented by the idiogram on the x-axis in the lower part of each figure.
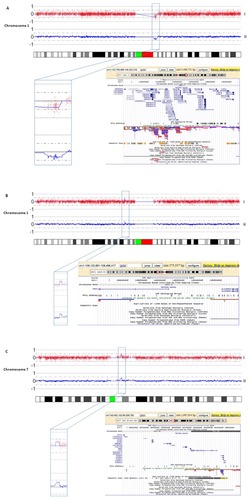
It remained unclear whether these CNVs had occurred de novo, since in none of the patients blood samples of the parents were available for carrier testing. A concise description of psychiatric and somatic phenotypes as well as ICD-10 classification is presented in .
Table 2 Phenotype of the three patients with a potentially pathogenic CNV
Discussion
In three out of the 80 patients with psychotic disorders (3.75%), one loss and two gains > 250 kb were detected which is in accordance with the report by Kirov who calculated a collective percentage of 3% CNVs in schizophrenia.Citation15
Apart from the recent report by Maiti et al,Citation25 who found three de novo CNVs (two gains and one loss) in 7q11.21 in two families with monozygotic twins discordant for schizophrenia in which, however, the breakpoints of these CNVs do not overlap with those in patient C, there is virtually no literature on the clinical significance. We recently detected a larger, completely overlapping gain of 2.4 Mb in 7q11.21 in a prenatal sample and the healthy father of this fetus with congenital diaphragmatic hernia. This makes it less likely that this gain is of clinical relevance.
With respect to 1p13.3, only Ohtsuki et al reported on a possible association with the netrin G1 (NTNG1) gene, located at 1p13.3, in Japanese families.Citation26 This does not, however, apply to patient B, nor does it clarify his symptom profile. Its relevance is further challenged by the detection of an exactly similar gain in 1p13.3 (in addition to a pathogenic, de novo duplication in 10q) in an intellectually disabled patient without any psychiatric symptoms who inherited this gain from his healthy father (unpublished data).
Microdeletions in the 1q21 region have been reported in several genome-wide analyses in large populations.Citation27,Citation28 A possible relationship between 1q21 and psychotic symptoms was reported by three research groups.Citation29–Citation31 Maiti et al reported on two de novo CNVs in families with monozygotic twins discordant for schizophrenia.Citation25 Several genes are mapped on 1q21, such as the KCNN3,Citation32 NOTCH2NL,Citation25 and Connexin 40/50.Citation33 Associations of these specific genes with schizophrenia in affected patients so far remain equivocal.
The results from this relatively small study using genome-wide array analysis do not differ essentially from the reported large scale studies. It has to be stressed that the commonly applied diagnostic categories, derived from the ICD or Diagnostic and Statistical Manual of Mental Disorders systems, show no relationship to a specific genetic etiology. Moreover, a single gene defect never codes for a categorical psychiatric disorder, but may lead to a biological dysfunction that has a certain probability to be associated with the development of an array of psychiatric symptoms.Citation14,Citation17,Citation18 This can best be illustrated by the discovery of the DISC1 gene in one family that was originally thought to be causally related to schizophrenia,Citation34 but later appeared to be involved in a biochemical cascade with consequences for neuronal functions predisposing for psychiatric symptoms across diagnostic boundaries.Citation35,Citation36
Although the CNVs detected in three out of 80 patients are not likely to be primarily involved in the evolution of their psychotic disorders, systematic genetic analysis in patients may reveal novel recurrent microdeletion syndromes that, however, are nearly always accompanied with developmental delay and dysmorphisms, and do not seem to correlate with psychiatric symptoms.Citation37,Citation38 The counterpart microduplications in these regions often cause a much milder phenotype.
In conclusion, the results of this and other studies on the presence of CNVs in patients with psychotic disorders have not yet led to a further specification of their pathophysiology nor to breakthroughs in clinical strategies. Therefore, while genetic analysis should always be considered as part of the diagnostic equipment in neuropsychiatry, the application of genome-wide array analysis in patients with psychotic disorders is mandatory only in the presence of other clinical signs such as facial dysmorphisms or developmental delay.
Acknowledgment
This study is part of a collaborative project of the research group ‘Psychopathology and Genetics’.
Disclosure
The authors report no conflicts of interest in this work.
References
- CuestaMJPeraltaVIntegrating psychopathological dimensions in functional psychoses: A hierarchical approachSchizophr Res20015221522911705715
- de GraciaDominguezViechtbauerWSimonsCPvan OsJKrabbendamLAre psychotic psychopathology and neurocognition orthogonal? A systematic review of their associationsPsychol Bull200913515717119210058
- McGrathJSahaSWelhamJEl SaadiOMacCauleyCChantDA systematic review of the incidence of schizophrenia: The distribution of rates and the influence of sex, urbanicity, migrant status and methodologyBMC Medicine20042131315115547
- Van OsJKapurSSchizophreniaLancet200937463564519700006
- SullivanPFThe genetics of schizophreniaPLoS Med20052e21216033310
- MortensenPBPedersenMGPedersenCBPsychiatric family history and schizophrenia risk in Denmark: Which mental disorders are relevant?Psychol Med20104020121019607751
- RutterMMoffittTECaspiAGene-environment interplay and psychopathology: Multiple varieties but real effectsJ Child Psychol Psychiatry20064722626116492258
- Van OsJKenisGRuttenBPFThe environment and schizophreniaNature201046820321221068828
- VeltmanJAGenomic microarrays in clinical diagnosisCurr Opin Pediatr20061859860317099357
- VissersLEde LigtJGilissenCA de novo paradigm for mental retardationNat Genet2010421109111221076407
- BassettASSchererSWBrzustowiczLMCopy number variations in schizophrenia: Critical review and new perspectives on concepts of genetics and diseaseAm J Psychiatry201016789991420439386
- DuanJSandersARGejmanPVGenome-wide approaches to schizophreniaBrain Res Bull2010839310220433910
- O’RoakBJDeriziotisPLeeCExome sequencing in sporadic autism spectrum identifies severe de novo mutationsNat Genet20114358558921572417
- VerhoevenWMAEggerJIMFeenstraIGenetic disorders and the autism spectrumClin Neuropsychiatry20118219224
- KirovGThe role of copy number variation in schizophreniaExpert Rev Neurother201010253220021318
- ItsaraACooperGMBakeCPopulation analysis of large copy number variants and hotspots of human genetic diseaseAm J Hum Genet20098414816119166990
- VermeeschJRBalikovaISchrander-StumpelCThe causality of de novo copy number variants is overestimatedEur J Hum Genet2011191112111321587321
- ArguelloPAGogosJAGenetic and cognitive windows into circuit mechanisms of psychiatric diseaseTrends Neurosci20123531322177981
- AndreasenNCFlaumMCArndtSThe comprehensive assessment of symptoms and history (CASH): An instrument for assessing diagnosis and psychopathologyArch Gen Psychiatry1992496156231637251
- KaySRFiszbeinAOpferLAThe Positive and Negative Syndrome Scale (PANSS) for schizophreniaSchizophr Bull1987132612763616518
- GuyWECDEU Assessment Manual for Psychopharmacology Revised NIMH Publication DHEW Publ. NO (Adm) 76-338Bethesda, MDNational Institute of Mental Health1976217222
- SkodolAERosnickKellmanDDevelopment of a procedure for validating structured assessment of Axis IIOldhamJMPersonality Disorders: New Perspectives on Diagnostic ValidityWashington, DCAmerican Psychiatric Press19914370
- World Health OrganizationThe ICD-10 Classification of Mental and Behavioural DisordersGeneva, SwitzerlandWorld Health Organization1992
- De LeeuwNHehir-KwaJYSimonsASNP array analysis in constitutional and cancer genome diagnostics – copy number variants, genotyping and quality controlCytogenet Genome Res201113521222121934286
- MaitiSKumarKHCastellaniCAO’ReillyRSinghSMOntogentic de novo copy number variations (CNVs) as a source of genetic individuality: studies on two families with MDZ twins for schizophreniaPlos One20116e1712521399695
- OhtsukiTHoriuchiYKogaMAssociation of polymorphisms in the haplotype block spanning the alternatively spliced exons of the NTNG1 gene at 1p13.3 with schizophrenia in Japanese populationsNeurosci Lett200843519419718384956
- StefanssonHRujescuDCichonSLarge recurrent microdeletions associated with schizophreniaNature200845523223618668039
- StoneJLO’DonovanMCGurlingHRare chromosomal deletions and duplications increase risk of schizophreniaNature200845523724118668038
- FananásLFusterCGuillamatRMiróRChromosomal fragile site 1q21 in schizophrenic patientsAm J Psychiatry1997154716716
- BrzustowiczMHodgkinsonKAChowEWHonerWGBassettASLocation of a major susceptibility locus for familial schizophrenia on chromosome 1q21–q22Science200028867868210784452
- RosaAFananasLCuestaMJPeraltaVShamP1q21–q22 locus is associated with susceptibility to the reality-distortion syndrome of schizophrenia spectrum disordersAm J Med Genet200211451651812116186
- KöhlerMHirschbergBBondCTSmall-conductance, calcium-activated potassium channels from mammalian brainScience1996273170917148781233
- NiXValenteJAzevedoMAPatoMTPatoCNKennedyJLConnexin 50 gene on human chromosome 1q21 is associated with schizophrenia in matched case-control and family-based studiesJ Med Genet20074453253617412882
- St ClairDBlackwoodDMuirWAssociation within a family of a balanced autosomal translocation with major mental illnessLancet199033613161973210
- MillarJKJamesRBrandonNJThomsonPADISC1 and DISC2: Discovering and dissecting molecular mechanisms underlying psychiatric illnessAnn Med20043636737815478311
- VerhoevenWMATuinierSClinical perspectives on the genetics of schizophrenia: A bottom-up orientationNeurotox Res20081414115019073422
- KoolenDASharpAJHurtsJAClinical and molecular delineation of the 17q21.31 microdeletion syndromeJ Med Genet20084571072018628315
- Van BonBWMMeffordHCMentenBFurther delineation of the 15q13 microdeletion and duplication syndromes: A clinical spectrum varying from non-pathogenic to a severe outcomeJ Med Genet20094651152319372089
- Hehir-KwaJEgmont-PetersenMJanssenIGenome-wide copy number profiling on high-density bacterial artificial chromosomes, single-nucleotide polymorphisms, and oligonucleotide microarrays: a platform comparison based on statistical power analysisDNA Res20071411117363414
