Abstract
Introduction
GRHL1 belongs to the family of Grainyhead-like (GRHL). Previous studies have shown that dysregulation of growth and survival pathways is associated with the GRHL family of gene cancers. Immunotherapy with checkpoint inhibitors has changed the treatment paradigm for many tumors, including endometrial cancer (EC). However, the effect of GRHL1 on immunotherapy in EC and its relationship with immune cell infiltration are poorly understood.
Methods
Differential expression of GRHL1 between EC and normal EC tissues was analyzed by searching the TCGA database, and the results were verified utilizing immunohistochemistry analyses. Next, the relationship between GRHL1, CD8+ T cells and tumor microenvironment (TME) was also investigated, and the effect of GRHL1 expression on immunotherapy in EC was evaluated.
Results
According to the findings, EC tissues had elevated expression levels of GRHL1 relative to normal tissues. Patients with EC who expressed GRHL1 at high levels experienced worse overall survival (OS) and Progression-free survival (PFS) than those whose expression was lower. In addition, GRHL1 expression was negatively correlated with CD8+ T cells, and patients with high GRHL1 expression were less effective in receiving immunotherapy.
Conclusion
The expression of GRHL1 was high in EC patients, and high expression of GRHL1 inhibits the proliferation of CD8+ T cells in the tumor microenvironment of EC and affect the efficacy of immunotherapy.
Introduction
Endometrial cancer (EC) is the sixth most common neoplasm in women worldwide.Citation1 In the US, EC is the most prevalent type of gynecological cancer.Citation2 In the US, the mortality rate for people of the white race was 4.3–4.5 per 100,000 individuals, while this value for people of the black race was 8.2–8.9 per 100,000 population. In China, the mortality rate for people of the yellow race was 2.5 per 100,000 individuals.Citation3 Even though high-grade EC often has a positive prognosis, it tends to recur. Because recurrent EC has such a dismal prognosis, preventing it is crucial. Currently, traditional surgery and minimally invasive surgery are the two most crucial treatment options for EC.Citation4,Citation5 The use of radiotherapy, immunotherapy, targeted therapy, and chemotherapy in the treatment of EC has increased in recent years.Citation6–8
The GRHL family of genes has been implicated in the development of many cancers,Citation9–13 and is involved in the regulation of embryogenesis, and growth and survival pathways in cancer,Citation14 where it leads to dysfunction through dedifferentiation or loss of functional integrity.Citation15 As a member of the GRHL family, GRHL1 is mainly associated with neuroblastoma and esophageal cancer.Citation16,Citation17
In this study, the possible involvement of GRHL1 in EC and its expression in TME were validated. Using the TCGA database and in vitro tests, we discovered that GRHL1 expression was considerably upregulated in EC tissues. We also explored the level of GRHL1 expression and its relationship with prognosis. In addition, the correlation of GRHL1 with the tumor microenvironment and CD8+ T cells was analyzed, and the effect of GRHL1 expression on EC immunotherapy was evaluated.
Materials and Methods
Dataset Source and Pre-Processing
The TCGA database was searched to retrieve clinical information on patients with uterine corpus endometrial carcinoma (UCEC), covering information on common gene expression, total mortality, and prognosis.Citation18 33 TCGA pan-cancer tumor mutation burden (TMB) and microsatellite instability (MSI) data downloaded from the UCLA Xena Data Portal (https://xenabrowser.net/). RNA sequencing (FPKM values) was downloaded from the TCGA Genome Data Commons (GDC, https://portal.gdc.cancer.gov/) and then scrutinized utilizing the R package TCGAbiolink.Citation18 The FPKM values were converted to transcript per kilobase million values. To account for the non-biological technical bias-related batch effect, the “ComBat” method from the sva package was implemented. R (v 4.1.2) and the R Bioconductor package were applied to analyze all of the data.
Clinical Sample Collection
Patients undergoing EC surgery at the First Affiliated Hospital of Bengbu Medical College, Department of Oncology Gynecology, from January 2021 to December 2021, were recruited for this research, and samples were obtained from those patients. Immunohistochemistry (IHC) labeling was performed on 66 EC tissue specimens and 10 surrounding normal tissues. Before or after surgery, no patient had chemotherapy, radiation, or biological treatment, and there was no history of EC in any of the patients. Tissue samples collected after surgery were preserved in a refrigerator at −80 degrees Celsius until protein extraction.
Experimental Materials
CUSABIO (CSB-PA868368LA01HU) provided rabbit anti-human antibody GRHL1 (50μL, WUHAN, CH). Primary antibodies against actin and alpha rabbit monoclonal antibodies against CD8+ T cells were purchased from Cell Signaling Technology Inc. (Danvers, MA, United States). Jackson ImmunoResearch Inc. provided an anti-rabbit antibody coupled to horseradish peroxidase (HRP, West Grove, PA, US). Sigma-Aldrich provided bovine serum albumin (BSA, St. Louis, MO, United States). Skim milk and Tween-20 were supplied by Sangon Biotech Co., Ltd (Shanghai, China).
Immunohistochemistry (IHC)
Paraformaldehyde (4%) (PFA) was employed to fix all tissue samples, followed by embedding them in paraffin, cutting them into sections, and attaching them on slides. Extraction of antigens was performed by soaking the slides in citrate buffer (pH 7.8, 0.1M) for 24 mins at about 82°C after they had been deparaffinized, rehydrated, and exposed to xylene density gradients. To inhibit peroxidase activity, slides were coated uniformly with an endogenous blocking solution for 15 min at room temperature (RT). Overnight, slides were treated with anti-GRHL1 primary antibody, then rinsed gently in PBS. Following a 10-minute incubation at RT with a biotin-conjugated secondary antibody, the samples were treated with streptavidin peroxidase for 5 mins. Next, the slides were rinsed with hematoxylin dye to eliminate the remaining debris. After the slides had been dried and washed, an IHC examination was performed. Sections were scored by two pathologists with extensive experience in double-blind reading. To determine the extent of staining, a quantity score (0–4) denoted 0, 0%; 1, 1–10%; 2, 11–50%; 3, 51–80% and 4, 81–100% of positive cells, was used. The staining intensity was divided into three grades: weak, moderate, and strong staining. It should be noted that the corresponding intensity scores ranged from 1 to 3. The final IHC score was calculated by multiplying the quantity and intensity scores.
Assessment of the Immune Characteristics of the EC Tumor Microenvironment
Immunological features of TME in EC encompass the immunomodulator expression, the cancer immune cycle activity, TIIC infiltration levels, and suppressive immune checkpoint expression. Information on 48 immunomodulators was collected, including chemokines and their receptors.Citation19 To examine the link between GRHL1 expression and the infiltration levels of immune cells, we employed the Tumour Immune Estimation Resource (TIMER)Citation20 and CIBERSORT algorithm to investigate how GRHL1 expression relates to immune cell activity. When the p-value was <0.05, it was considered to be significant. To avoid errors, correlations between GRHL1 and CD8+ T cells were calculated based on 7 different algorithms: XCELL, QUANTISEQ, CIBERSORT−ABS, CIBERSORT, MCP−COUNTER, TIMER and EPIC algorithms.Citation21–27
Immune Response Analysis
The Cancer Genome Atlas (TCGA) and other sources of next-generation sequencing (NGS) data for 20 solid tumors are provided with an extensive immunogenomic analysis by the Cancer Immunome Database (TCIA) (https://www.tcia.at/home).Citation28 Using the ggpubr R program, the immunological phenomenon scores (IPS) of 560 EC patients from this database were used to assess immunotherapy.
Statistical Analysis
R software (version 4.1.2) and Perl (version 5.32.1.1) were utilized for data analysis. P <0.05 indicated the significance level.
Results
Expression of GRHL1 in Endometrial Cancer and Adjacent Normal Tissues and Its Prognostic Assessment in Endometrial Cancer
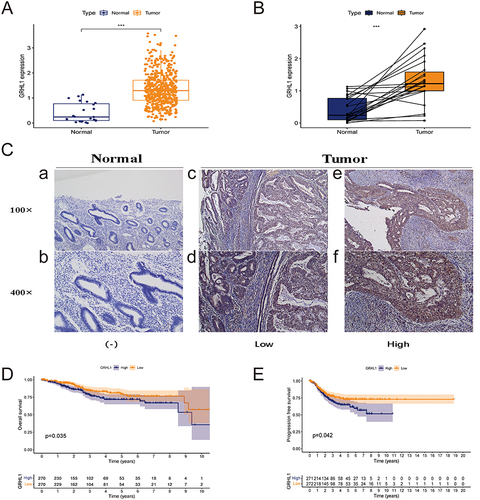
In the TCGA cohort, the GRHL1 mRNA expression level was found to be elevated in EC tissues compared to nearby normal tissues (p < 0.001, ). The comparison of the mRNA expression of GRHL1 in 35 pairs of EC and adjacent normal tissues in the TCGA database showed that GRHL1 expression was higher in EC tissues than in adjacent normal tissues (p < 0.05, ). IHC staining tests confirmed this finding at the tissue level (). KM analysis of survival of patients exhibiting varying GRHL1 expression levels based on TCGA-derived data was performed to evaluate the prognostic significance of GRHL1 in EC. According to the results, the 10-year OS was substantially higher in patients whose GRHL1 expression was lower as compared to those whose GRHL1 expression was higher (p =0.035, ). We also discovered that GRHL1 overexpression was a poor predictor of EC PFS (p = 0.042, ). Overall, remarkably higher expression levels of GRHL1 were found in EC, in comparison to nearby normal tissues. GRHL1 upregulation was associated with a shorter OS, and PFS for patients.
Figure 1 Expression of GRHL1 in endometrial cancer and adjacent normal tissues and its prognostic assessment in endometrial cancer. (A) GRHL1 was overexpressed in the TCGA cohort. (B) In the TCGA cohort, GRHL1 was over-expressed in the tumor relative to the noncancerous tissues. (C) IHC staining analysis showed that GRHL1 expression was low in paracancerous tissues (a and b) and up-regulated in EC tissues (c–f). (D) Kaplan-Meier analysis of OS of individuals with EC with GRHL1 gene expression in the TCGA database. (E) Kaplan-Meier analysis of PFS of individuals with EC with GRHL1 gene expression in the TCGA database. (***p < 0.001).
Clinicopathological and Predictive Value of GRHL1 Expression in EC Patients
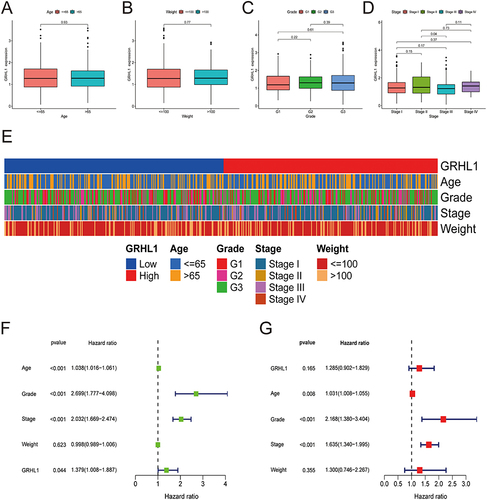
Using box plots and heatmaps, we revealed that GRHL1 expression levels were correlated with clinicopathological parameters of EC patients (). The results showed that the expression level of GRHL1 did not correlate well with the clinicopathologic stage. Patients’ clinical stage, grade, and age were shown to have a substantial association with their OS, and GRHL1 was found to have statistically significant predictive value for EC patients, as shown in a univariate analysis (p = 0.044, ). In addition, multivariate analysis demonstrated that clinical stage, grade, and age were substantially linked to EC patients’ OS, but the prognostic value of GRHL1 for EC patients was found to be insignificant (p = 0.165, ).
Figure 2 Association of clinicopathological characteristics with GRHL1 expression levels. (A–D) The association of GRHL1 expression level with the clinicopathological features of individuals with EC for age (A), weight (B), Grade (C), and stage (D). (E) Heatmap illustrating the link between the clinicopathological characteristics of EC patients and the GRHL1 expression. (F and G) Forest plots show the results of univariate (F) and multivariate (G) Cox regression analyses for the OS of patients with EC.
GRHL1 Inhibits CD8+ T Cell Proliferation and is Associated with Immune Escape in EC
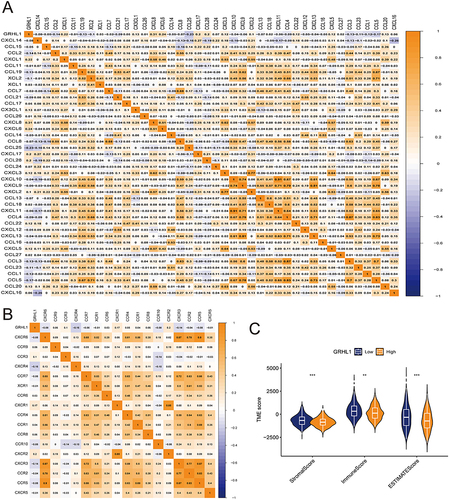
Herein, we assessed the correlation of chemokines and their receptors with GRHL1 expression ( and ). We found that CCL2, CCL4, CCL4, CCL5, CXCL9, CXCL10, CXCR3, and CCR5 were negatively correlated with the expression of GRHL1. Chemokines that bind to CXCR3 (such as CXCL9 and CXCL10) are essential and necessary for the trafficking of activated CD8+ T cells to tumor sites.Citation29 Subsequently, we used the TCGA-derived data to calculate the TME scores using the EC expressions and the “estimate” package included R. Furthermore, we plotted TME differential analysis. The ImmuneScore, StromalScore, and ESTIMATEScore of TME were downregulated in the high GRHL1 expression group ().
Figure 3 Correlation of GRHL1 with chemokines and tumor microenvironment in EC. (A and B) Correlation of GRHL1 with the immunomodulator chemokine (A) and its receptor (B) in EC. (C) The TME’s ESTIMATEScore, ImmuneScore, and StromalScore varied between the groups with high and low GRHL1 expression. (**p < 0.01; ***p < 0.001).
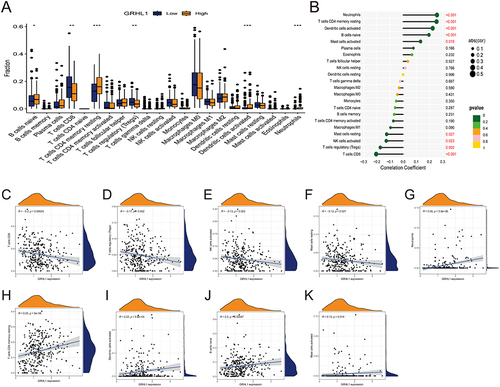
The CIBERSORT algorithm was used to verify the correlation between GRHL1 and immune cells (). The abundance of 22 different types of immune cells was measured and compared between the two groups (). GRHL1 was negatively linked to CD8+ T cells, T cells regulatory (Tregs), NK cells activated, and Mast cells resting, and positively correlated with Neutrophils, T cells CD4 memory resting, Dendritic cells activated, B cells naive, and Mast cells activated (). In previous studies, TME has been categorized into two subtypes as follows: an inflammatory TME dominated by T-cell infiltration and a non-inflammatory TME dominated by T-cell suppression. Tumors with T-cell inflammation contain abundant CD8+ T cells. Tumors without T-cell inflammation lack these cells but contain blood vessels, fibroblasts, and macrophages, which support tumor growth.Citation30,Citation31
Figure 4 Link between GRHL1 and immune cell infiltration. (A) Box plots showing the differences between 22 immune cell types and GRHL1 expression analysis in EC. (B) Correlation between GRHL1 expression and 22 immune cell types in EC. (C–F) GRHL1 expression was negatively linked to CD8+ T cells (C), T cells regulatory (Tregs) (D), NK cells activated (E), Mast cells resting (F). (G–K) GRHL1 expression was positively correlated with Neutrophils (G), T cells CD4 memory resting (H), Dendritic cells activated (I), B cells naïve (J), and Mast cells activated (K). (*p < 0.05; **p <0.01; ***p < 0.001).
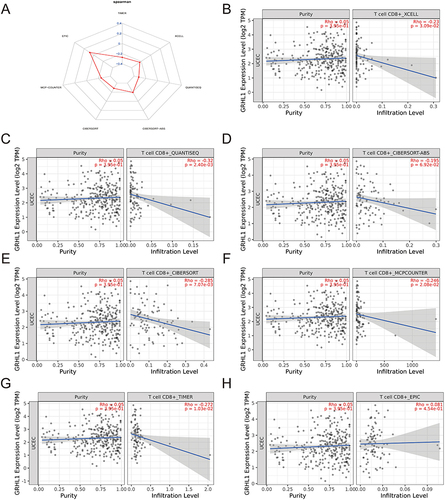
Next, we showed GRHL1 correlation with CD8+ T cells based on ssGSEA, MCPcounter, TIMER, ESTIMATE, CIBERSORT, XCELL, and EPIC algorithms (). Among the six algorithms, GRHL1 correlated negatively with CD8+ T cells (). TME cell infiltration is characterized by three types: immunoinflammatory phenotype (immunoinflammatory), immune exclusion phenotype (immune), and immune desert. The immune exclusion and the immune desert phenotypes are also known as non-immune inflammatory phenotypes.Citation32 Taken together, we define the immune microenvironment shaped by GRHL1 as an immunodeficient phenotype (immune exclusion phenotype), that is, a non-immune inflammatory phenotype.
Figure 5 Investigation of the relationship between GRHL1 expression and CD8+ T cells. (A) Radar plot of correlation analysis between GRHL1 expression and CD8+ T cells. (B–H) Seven immunological algorithms were utilized to investigate the link between GRHL1 expression and CD8+ T cells: XCELL (B), QUANTISEQ (C), CIBERSORT−ABS (D), CIBERSORT (E), MCP−COUNTER (F), TIMER (G) and EPIC algorithms (H).
Analyses of Immune Cell Infiltration in High- and Low-GRHL1-Expression Groups
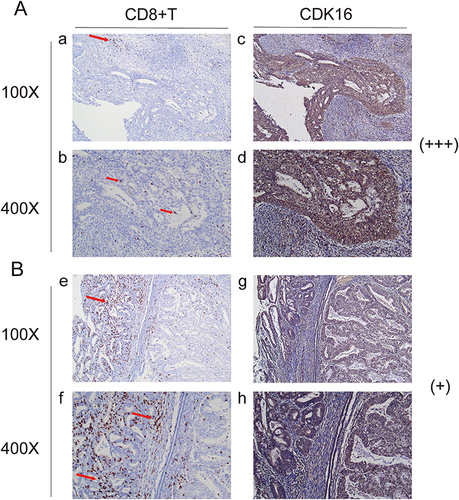
Since GRHL1 was negatively correlated with CD8+ T cells, we investigated its potential role in the tumor immune microenvironment (TIME) of EC. IHC staining was performed to compare the tissue samples with low (+) and high (+++) GRHL1 expression levels (). To determine whether there was a link between high GRHL1 expression and CD8+ T cells in TIME, we used IHC labeling to measure the infiltration of CD8+ T cells into EC and the surrounding tissues. CD8+ T cells in EC tissues with high (+++) GRHL1 levels were particularly clustered in the peripheral tumor stroma, with just a small proportion of immune cells infiltrating the stroma to reach the tumor’s parenchyma (). Conversely, a greater number of immune cells infiltrated the stroma of the tumor parenchyma, and fewer CD8+ T cells were clustered throughout the tumor parenchyma in EC tissues with low (+) GRHL1 levels (). In summary, high expression of GRHL1 inhibited the proliferation of CD8+ T cells in the TIME of EC, as shown by the analysis.
Figure 6 Immune cell infiltration in the groups with high and low GRHL1 expression. (A) CD8+ T cell (a and b) infiltration in the group with high GRHL1 expression (c and d). (B) CD8+ T cell (e and f) infiltration in the group with low GRHL1 expression (g, h). The locations of immunological cells are indicated by the red arrows.
Correlation Analysis of GRHL1 Expression with TMB and MSI and Evaluation of Immunotherapy
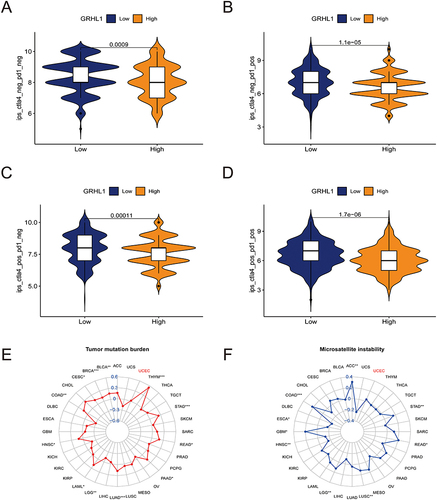
After exploring immune phenomenon scores (IPS) through TCIA,Citation28 it was observed that patients with high GRHL1 expression were less effective in receiving immunotherapy (, p < 0. 05). To exclude the influence of confounding factors on immunotherapy efficacy, we also assessed the correlation between TMB/MSI and GRHL1 expression. ( and , p > 0. 05).
Figure 7 Correlation analysis of GRHL1 expression with TMB and MSI and evaluation of immunotherapy. (A–D) Association of IPS with the GRHL1 expression in individuals with EC based on the TCIA database; CTLA4- PD1- (A), CTLA4- PD1+ (B), CTLA4+ PD1- (C), CTLA4+PD1+ (D). (E and F) Correlation analysis of GRHL1 expression with TMB (E) and MSI (F). (*p < 0.05; **p <0.01; ***p < 0.001).
Discussion
Endometrial cancer (EC) is among the three most prevalent gynecological malignancies.Citation33 It has now become the most common gynecologic cancer in developed countries.Citation33 Although the majority of patients are diagnosed at an early stage and have a favorable prognosis, there are still some patients with advanced stage at the time of initial diagnosis or recurrence and metastasis after treatment, and the 5-year survival rate is only 20–26%.Citation34 The success of tumor immunotherapy has shown that immunotherapy plays a landmark role in cancer treatment.Citation35 In recent years, treatment guidelines for EC have been updated to include targeted treatments such as immune checkpoint inhibitors.Citation36,Citation37
Homologous sequencing has led to the identification of 3 GRHL genes in the human genome to date.Citation14 Recent years have seen a surge in the publication of multiple studies on GRHL and cancer, and these findings provide compelling evidence that GRHL is strongly associated with cancer.Citation15,Citation38,Citation39 GRHL3 is closely associated with epithelial cancers, including skin, breast, and head and neck cancers.Citation10,Citation11,Citation40 GRHL2 acts as an oncogene to suppress epithelial-to-mesenchymal transition (EMT).Citation41,Citation42 In recent years GRHL1 has also been found to be closely associated with skin cancer, esophageal cancer and neuroblastoma.Citation16,Citation17,Citation43
We discovered that GRHL1 expression was upregulated in EC tissues in comparison to non-EC tissues. GRHL1 upregulation was associated with a shorter OS, and PFS for patients. Based on these results, GRHL1 could be a potential target gene for EC. It has been shown that immune infiltration and immune escape of tumors correlate with both the prognosis of cancer and the patient’s response to treatment.Citation44 The majority of tumor cells express antigens that may facilitate the identification of the tumor by the host CD8+ T cells. The TME allows for the categorization of immune escape into two distinct subtypes. One of the primary subsets has a phenotype that is indicative of T cell inflammation. These tumors can resist immune attack through the dominant inhibitory effects of immune system-suppressive pathways. The other group does not exhibit this T cell inflammation profile and hence is resistant to immunological assault due to being excluded or ignored by the immune system.Citation45 Standardized cancer immunotherapy requires that a target molecule exhibit TME-specific upregulation and immunosuppression action.Citation46 To determine the potential role of GRHL1 in EC and its expression in TME, we explored the EC cohort from the transcriptome of specimens from the TCGA database dataset, and the results showed that EC expression was significantly overexpressed in tumor tissues. Additional research using IHC validated these findings. Our data also demonstrated a negative association between GRHL1 and the immunological states of TME in EC. Using seven different algorithms, we found that in the high GRHL1 group, T cell recruitment activity was remarkably downregulated, GRHL1 was negatively linked to CD8+ T cell activation, and the amount of TIIC infiltration was substantially decreased. These results suggest that high expression of GRHL1 in EC may inhibit the proliferation of CD8+ T cells, thereby affecting the efficacy of immunotherapy. Subsequent IHC experiments also verified this result. Therefore, targeting GRHL1 in EC may improve the success of immunotherapy. Previous studies have shown that about 30% of endometrial cancers have mismatch repair defects, which increase the adverse effects of drug therapy, while the defects increase the likelihood of a positive response to immune checkpoint inhibitors.Citation47 And there is growing evidence that tumor mutational load is a promising predictor of immune checkpoint inhibitor therapy.Citation48 To rule out the influence of the confounding factors TMB and MSI on the efficacy of immunotherapy, we assessed the correlation between TMB/MSI and GRHL1 expression. We found that there was no significant correlation between GRHL1 expression level and TMB/MSI in endometrial cancer. This result also verified our conclusion from other aspects.
Some limitations remain in this study. The TCGA data served as the foundation for our investigation of GRHL1’s involvement in EC. Despite our success in using IHC assays to verify the link between GRHL1 and immune cells infiltrating TME, the absence of equivalent confirmation in cellular and animal trials suggests where we should focus our future efforts. Second, the effect of GRHL1 on patient immunotherapy was derived from an analysis of the TCGA database and lacks direct evidence. Third, although we analyzed a direct link between GRHL1 expression and MSI, we did not stratify the mismatch repair status of the tumor on consecutive samples and TCGA data, and therefore could not obtain GRHL1 results for the mismatch repair good and mismatch repair bad subgroups, which is a direction for our future work.
Conclusion
Our results show that GRHL1 is highly expressed in EC tissues compared to normal samples and can be characterized as a specific biomarker to distinguish EC tissues from normal endometrial tissue. Unfavourable OS, and PFS were all highly correlated with GRHL1 upregulation in EC patients. Immune cell infiltration analysis showed that high expression of GRHL1 inhibited the proliferation of CD8+ T cells and affected the efficacy of immunotherapy. Our thorough analysis of GRHL1’s potential as an immune target for EC may offer a useful assessment system for clinical use.
Abbreviations
EC, endometrial cancer; GRHL1, Grainyhead like transcription factor 1; TCGA, The Cancer Genome Atlas; OS, overall survival; GSEA, Gene set enrichment analysis; IHC, Immunohistochemistry; RT, room temperature; DCs, resting dendritic cells; KM, Kaplan-Meier; FDR, false discovery rate; TME, tumor microenvironment; TIME, Tumor immune microenvironment; TMB, tumor mutation burden; ICI, Immune checkpoint inhibitor; ICB, immune checkpoint blockade; MSigDB, Molecular Signatures Database; FDRs, false discovery rates; TIMER, Tumour Immune Estimation Resource; TCIA, The Cancer Immunome Database; IPS, immune phenomenon scores.
Data Sharing Statement
The following online resources were screened based on this study’s analysis of used clinical data: TCGA (https://www.cancer.gov/), TCIA (https://tcia.at/).
Ethics Approval and Informed Consent
The study involving human participants has been reviewed and approved by the First Affiliated Hospital of Bengbu Medical College’s [2021] 143 ethics committee. The Helsinki Declaration, which outlines moral guidelines for medical research with human participants, was followed in conducting the study. Patients/participants signed a consent form to indicate their agreement to be included in the study with each patient providing their informed consent.
Author Contributions
All authors contributed significantly to the research that was published, whether it was in the conceptualization, research design, implementation, data gathering, analysis, and interpretation, or each of these areas separately; participated in the report’s drafting, revision, or detailed evaluation; approved the final version for publishing; decided upon the journal to which the manuscript has been submitted; and accept responsibility for all aspects of the project.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgments
We thank Bullet Edits Limited for their invaluable assistance with language editing and manuscript proofreading. In addition, we also acknowledge the open databases, including TCGA, and TCIA, for providing the necessary platform and datasets for this research.
Additional information
Funding
References
- Lortet-Tieulent J, Ferlay J, Bray F, Jemal A. International patterns and trends in endometrial cancer incidence, 1978–2013. J Natl Cancer Inst. 2018;110(4):354–361. doi:10.1093/jnci/djx214
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
- Huo X, Sun H, Liu Q, et al. Clinical and expression significance of AKT1 by co-expression network analysis in endometrial cancer. Front Oncol. 2019;9:1147. doi:10.3389/fonc.2019.01147
- Chambers LM, Carr C, Freeman L, Jernigan AM, Michener CM. Does surgical platform impact recurrence and survival? A study of utilization of multiport, single-port, and robotic-assisted laparoscopy in endometrial cancer surgery. Am J Obstet Gynecol. 2019;221(3):243 e241–243 e211. doi:10.1016/j.ajog.2019.04.038
- Wright JD, Khoury-Collado F, Melamed A. Harnessing minimally invasive surgery to improve outcomes in endometrial cancer surgery-the robots are coming. JAMA Surg. 2019;154(6):539. doi:10.1001/jamasurg.2018.5841
- Aoki Y, Kanao H, Wang X, et al. Adjuvant treatment of endometrial cancer today. Jpn J Clin Oncol. 2020;50(7):753–765. doi:10.1093/jjco/hyaa071
- Ott PA, Bang YJ, Berton-Rigaud D, et al. Safety and antitumor activity of pembrolizumab in advanced programmed death ligand 1-positive endometrial cancer: results from the KEYNOTE-028 Study. J Clin Oncol. 2017;35(22):2535–2541. doi:10.1200/JCO.2017.72.5952
- Makker V, Rasco D, Vogelzang NJ, et al. Lenvatinib plus pembrolizumab in patients with advanced endometrial cancer: an interim analysis of a multicentre, open-label, single-arm, Phase 2 trial. Lancet Oncol. 2019;20(5):711–718. doi:10.1016/S1470-2045(19)30020-8
- Darido C, Georgy SR, Wilanowski T, et al. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell. 2011;20(5):635–648. doi:10.1016/j.ccr.2011.10.014
- Georgy SR, Cangkrama M, Srivastava S, et al. Identification of a novel proto-oncogenic network in head and neck squamous cell carcinoma. J Natl Cancer Inst. 2015;107(9). doi:10.1093/jnci/djv152
- Bhandari A, Gordon W, Dizon D, et al. The Grainyhead transcription factor Grhl3/Get1 suppresses miR-21 expression and tumorigenesis in skin: modulation of the miR-21 target MSH2 by RNA-binding protein DND1. Oncogene. 2013;32(12):1497–1507. doi:10.1038/onc.2012.168
- Werner S, Frey S, Riethdorf S, et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J Biol Chem. 2013;288(32):22993–23008. doi:10.1074/jbc.M113.456293
- Quan Y, Xu M, Cui P, Ye M, Zhuang B, Min Z. Grainyhead-like 2 promotes tumor growth and is associated with poor prognosis in colorectal cancer. J Cancer. 2015;6(4):342–350. doi:10.7150/jca.10969
- Gasperoni JG, Fuller JN, Darido C, Wilanowski T, Dworkin S. Grainyhead-like (Grhl) target genes in development and cancer. Int J Mol Sci. 2022;23(5):2735. doi:10.3390/ijms23052735
- Kotarba G, Taracha-Wisniewska A, Wilanowski T. Grainyhead-like transcription factors in cancer - Focus on recent developments. Exp Biol Med. 2020;245(5):402–410. doi:10.1177/1535370220903009
- Fabian J, Lodrini M, Oehme I, et al. GRHL1 acts as tumor suppressor in neuroblastoma and is negatively regulated by MYCN and HDAC3. Cancer Res. 2014;74(9):2604–2616. doi:10.1158/0008-5472.CAN-13-1904
- Li M, Li Z, Guan X, Qin Y. Suppressor gene GRHL1 is associated with prognosis in patients with oesophageal squamous cell carcinoma. Oncol Lett. 2019;17(5):4313–4320. doi:10.3892/ol.2019.10072
- Colaprico A, Silva TC, Olsen C, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. doi:10.1093/nar/gkv1507
- Charoentong P, Finotello F, Angelova M, et al. Pan-cancer Immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi:10.1016/j.celrep.2016.12.019
- Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259.
- Xu L, Deng C, Pang B, et al. TIP: a web server for resolving tumor immunophenotype profiling. Cancer Res. 2018;78(23):6575–6580. doi:10.1158/0008-5472.CAN-18-0689
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi:10.1093/bioinformatics/btz210
- Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi:10.1093/nar/gkaa407
- Finotello F, Mayer C, Plattner C, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11(1):34. doi:10.1186/s13073-019-0638-6
- Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17(1):218. doi:10.1186/s13059-016-1070-5
- Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi:10.1186/s13059-016-1028-7
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
- Prior FW, Clark K, Commean P, et al. TCIA: an information resource to enable open science. Annu Int Conf IEEE Eng Med Biol Soc. 2013;2013:1282–1285. doi:10.1109/EMBC.2013.6609742
- Mikucki ME, Fisher DT, Matsuzaki J, et al. Non-redundant requirement for CXCR3 signalling during tumoricidal T-cell trafficking across tumour vascular checkpoints. Nat Commun. 2015;6(1):7458. doi:10.1038/ncomms8458
- Gajewski TF, Corrales L, Williams J, Horton B, Sivan A, Spranger S. Cancer immunotherapy targets based on understanding the T cell-inflamed versus non-T cell-inflamed tumor microenvironment. Adv Exp Med Biol. 2017;1036:19–31.
- Garris CS, Luke JJ. Dendritic cells, the T-cell-inflamed tumor microenvironment, and immunotherapy treatment response. Clin Cancer Res. 2020;26(15):3901–3907. doi:10.1158/1078-0432.CCR-19-1321
- Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19(1):53. doi:10.1186/s12943-020-01170-0
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
- Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet. 2016;387(10023):1094–1108. doi:10.1016/S0140-6736(15)00130-0
- Couzin-Frankel J. Breakthrough of the year 2013. Cancer immunotherapy. Science. 2013;342(6165):1432–1433. doi:10.1126/science.342.6165.1432
- Chambers SK. Advances in chemotherapy and targeted therapies in endometrial cancer. Cancers. 2022;14(20):5020. doi:10.3390/cancers14205020
- Mahdi H, Chelariu-Raicu A, Slomovitz BM. Immunotherapy in endometrial cancer. Int J Gynecol Cancer. 2023;33(3):351–357. doi:10.1136/ijgc-2022-003675
- Mathiyalagan N, Miles LB, Anderson PJ, et al. Meta-analysis of grainyhead-like dependent transcriptional networks: a roadmap for identifying novel conserved genetic pathways. Genes. 2019;10(11):876. doi:10.3390/genes10110876
- Goldie SJ, Cottle DL, Tan FH, et al. Loss of GRHL3 leads to TARC/CCL17-mediated keratinocyte proliferation in the epidermis. Cell Death Dis. 2018;9(11):1072. doi:10.1038/s41419-018-0901-6
- Xu H, Liu C, Zhao Z, et al. Clinical implications of GRHL3 protein expression in breast cancer. Tumour Biol. 2014;35(3):1827–1831. doi:10.1007/s13277-013-1244-7
- Pifer PM, Farris JC, Thomas AL, et al. Grainyhead-like 2 inhibits the coactivator p300, suppressing tubulogenesis and the epithelial-mesenchymal transition. Mol Biol Cell. 2016;27(15):2479–2492. doi:10.1091/mbc.e16-04-0249
- Chung VY, Tan TZ, Tan M, et al. GRHL2-miR-200-ZEB1 maintains the epithelial status of ovarian cancer through transcriptional regulation and histone modification. Sci Rep. 2016;6(1):19943. doi:10.1038/srep19943
- Mlacki M, Darido C, Jane SM, Wilanowski T. Loss of Grainy head-like 1 is associated with disruption of the epidermal barrier and squamous cell carcinoma of the skin. PLoS One. 2014;9(2):e89247. doi:10.1371/journal.pone.0089247
- Jhunjhunwala S, Hammer C, Delamarre L Antigen presentation in cancer: insights into tumour immunogenicity and immune evasion. Nat Rev Cancer. 2021;21(5):298–312. doi:10.1038/s41568-021-00339-z
- Kaderbhai C, Tharin Z, Ghiringhelli F. The role of molecular profiling to predict the response to immune checkpoint inhibitors in lung cancer. Cancers. 2019;11(2):201. doi:10.3390/cancers11020201
- Sanmamed MF, Chen L. A paradigm shift in cancer immunotherapy: from enhancement to normalization. Cell. 2018;175(2):313–326. doi:10.1016/j.cell.2018.09.035
- Yang Z, Wei S, Deng Y, Wang Z, Liu L. Clinical significance of tumour mutation burden in immunotherapy across multiple cancer types: an individual meta-analysis. Jpn J Clin Oncol. 2020;50(9):1023–1031. doi:10.1093/jjco/hyaa076
- Talhouk A, McConechy MK, Leung S, et al. A clinically applicable molecular-based classification for endometrial cancers. Br J Cancer. 2015;113(2):299–310. doi:10.1038/bjc.2015.190
