To the Editor,
Histone acetylation in conjunction with deacetylation serves as a crucial modulation process of chromatin structure and is one of the main epigenetic mechanisms for gene regulation [Citation1]. The status of histone acetylation is controlled by histone acetyltransferases and histone deacetylase (HDAC). HDACs are responsible for the deacetylation of lysine residues on the N-terminal region of the core histones (H2A, H2B, H3 and H4). Human HDACs consist of 11 proteins in four families (class I: HDAC1, 2, 3 and 8; class IIa: HDAC4, 5, 7 and 9; class IIb: HDAC6 and 10; class IV: HDAC11) [Citation1]. HDAC-mediated deacetylation alters activities of many transcription factors, including p53, nuclear factor κB, and HIF1α, subsequently regulating many cellular events, including organ development, angiogenesis, apoptosis and cell proliferation [Citation1].
High expression of HDACs is a common feature of cancer cells, and HDACs are considered promising targets for cancer therapy [Citation2]. However, HDAC inhibitor resistance has been reported in cancer cells [Citation2]. One of the mechanisms for the resistance is inactivation of HDAC2 by frameshift mutation in HDAC2 gene, which preferentially occurs in cancers with microsatellite instability (MSI) (up to 29% in the cancers with MSI) [Citation3]. Frameshift mutations of genes containing mononucleotide repeats are features of gastric (GC), colorectal carcinomas (CRC) and endometrial carcinomas with MSI [Citation4]. In addition to HDAC2, we found mononucleotide repeats in the coding sequences of HDAC4, 5, 6, 7, 9 and 11 genes (http://genome.cse.ucsc.edu/), but the frameshift mutations at these sites are currently unknown.
To see whether the mononucleotide repeats in HDAC4, 5, 6, 7, 9 and 11 genes are mutated in GC and CRC with MSI, we analyzed exon 22 of HDAC4 (C7), exon 22 of HDAC5 (C7), exon 25 of HDAC6 (C7), exon 21 of HDAC7 (C7), exon 15 of HDAC9 (T7) and exon 4 of HDAC11 (C7) by polymerase chain reaction (PCR)-based single strand conformation polymorphism (SSCP) assay. For the mutation analysis, methacarn-fixed tissues of 42 GC and 51 CRC with MSI were used in this study. These cancers consisted of 30 GC with high MSI (MSI-H), 12 GC with low MSI (MSI-L), 36 CRC with MSI-H and 15 CRC with MSI-L. Malignant cells and normal cells from the same patients were selectively procured from hematoxylin and eosin-stained slides using a 30G1/2 hypodermic needle affixed to a micromanipulator [Citation5]. Genomic DNA each from tumor cells and corresponding normal cells were amplified with specific primer pairs by PCR. Radioisotope ([32P]dCTP) was incorporated into the PCR products for detection by SSCP autoradiogram. After SSCP, migration of the PCR products on the SSCP was analyzed by visual inspection. Direct DNA sequencing reactions were performed in the cancers with the mobility shifts in the SSCP. Other procedures of the PCR and SSCP were described in our previous studies [Citation5].
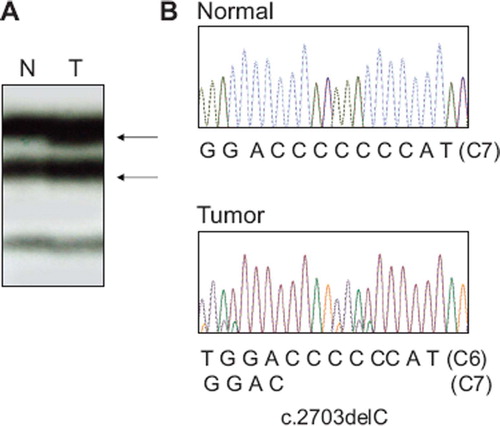
On the SSCP, all of the PCR products from the cancers were clearly seen. However, there was only one cancer showing aberrant bands on SSCP. PCR-SSCP analysis of the HDAC4 exon 22 identified aberrant bands in one (3.3%) of the 30 GC with MSI-H (). In this case, DNA from normal tissues from the same patients with the HDAC4 mutation showed no evidence of the mutation in SSCP, indicating the mutations had risen somatically. Direct DNA sequencing analysis of the GC with the aberrant SSCP bands led to identification of a HDAC4 deletion mutation within the C7 repeat sequences (). The mutation was c.2703delC, which would result in a premature stop of the amino acid synthesis (p.Met902TrpfsX55). There was no somatic mutation of HDAC5 or HDAC6 or HDAC7 or HDAC9 or HDAC11 in the cancers analyzed. We repeated the experiments twice, including PCR, SSCP and DNA sequencing analysis to ensure the specificity of the results, and found that the data were consistent. We also analyzed PIK3CA gene mutation, a prevalent mutation, in the same GC samples by the SSCP. We detected three PIK3CA mutations (3/40; 8%) the frequency of which was similar to its known frequency in GC [Citation5]. Together, these data show that the mutational analysis was sensitive to detect the mutations.
Figure 1. Mutation of HDAC4 exon 22 in a gastric carcinoma with MSI-H. A. PCR product of HDAC4 exon 22 from a gastric carcinoma shows aberrant bands (arrows in lane T) as compared to SSCP from normal tissue (N) of the same patient. B. Direct DNA sequencing analysis shows a heterozygous C deletion within the C7 in tumor tissue as compare to normal tissue.
Although HDAC isoforms reveal specificity in some functional aspects, they show redundancy in other aspects [Citation1,Citation2]. Also, HDAC inhibitors are widely effective in inhibiting HDAC isoforms. Because it is possible that inactivation of HDAC isoforms cooperatively contribute to cancer development and HDAC inhibitor resistance, we analyzed frameshift mutation of HDAC4, 5, 6, 7, 9 and 11 genes at the mononucleotide repeats. However, we detected only one mutation in the cancers with MSI, suggesting that the frameshift mutations of the HDAC genes besides HDAC2 are rare in GC and CRC with MSI, and that they may not play a crucial role in the cancer development or the therapy resistance. However, it remains unknown that other genetic alterations of HDAC genes besides the frameshift mutation are present in human cancers. To see whether genetic alteration of HDAC genes is a common feature of human cancers, the status of HDAC genes should be further analyzed at various levels in many cancer types.
Acknowledgements
This study was supported by a grant from National Research Foundation of Korea (R01-2008-000-10014-0).
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nat Rev Genet 2009; 10:32–42.
- Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol 2009;27:5459–68.
- Ropero S, Fraga MF, Ballestar E, Hamelin R, Yamamoto H, Boix-Chornet M, . A truncating mutation of HDAC2 in human cancers confers resistance to histone deacetylase inhibition. Nat Genet 2006;38:566–9.
- Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: The interrelationship between genetics and epigenetics. Carcinogenesis 2008;29:673–80.
- Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, . PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene 2005;24: 1477–80.
