Abstract
Patient outcome from glioma may be influenced by germline variation. Considering the importance of DNA repair in cancer biology as well as in response to treatment, we studied the relationship between 1458 SNPs, which captured the majority of the common genetic variation in 136 DNA repair genes, in 138 glioblastoma samples from Sweden and Denmark. We confirmed our findings in an independent cohort of 121 glioblastoma patients from the UK. Our analysis revealed nine SNPs annotating MSH2, RAD51L1 and RECQL4 that were significantly (p < 0.05) associated with glioblastoma survival.
Glioblastoma, the most aggressive type of primary brain tumour, is associated with a very poor prognosis. Well-established positive prognostic factors include young age at diagnosis, high performance status, O(6)-methylguanine methyltransferase (MGMT) promoter methylation, and concomitant chemo and radiation treatment. Although these prognostic factors are useful for predicting prognosis and treatment requirements, there is variability in clinical outcome for patients with apparently the same stage disease. Hence accurate assessment of prognosis would be beneficial when choosing specific therapeutic options and an assessment of the likelihood of response and adverse reactions to chemotherapeutic treatments would allow patient-tailored decisions on drug selection. These assessments could improve survival. In addition to somatic differences, germline variation probably also plays a role in defining individual patient outcome.
Conventional glioblastoma treatment, with radio- and chemotherapy, aims to inhibit cancer cells by promoting DNA damage. In turn, the cancer cells’ sensitivity to treatment partly depends on their ability to repair the sustained DNA damage [Citation1]. Therefore, DNA repair genes involved in restoring DNA after ionising radiation and chemotherapy are central genes to study in association with glioblastoma outcome. Aside from homologous recombination (a key mechanism that repairs radiation-induced DNA double strand breaks), there are specific mechanisms for nucleotide excision repair (NER) as well as DNA mismatch repair. Mutations in the repair pathways typically sensitise cells towards radiotherapy. For example, inhibition of RAD51 family members, key components in repairing double strand breaks through homologous recombination, enhances the radioresistance of glioma cells [Citation2].
We have comprehensively investigated a set of 1127 tag SNPs and 388 putative functional SNPs, all mapped to DNA-repair genes, in association with glioblastoma outcome. Through a collaborative international study, we have collected and genotyped 172 glioblastoma samples from Sweden and Denmark and confirmed significant findings in an independent dataset of 295 glioblastoma cases from northern UK.
Material and methods
Patients
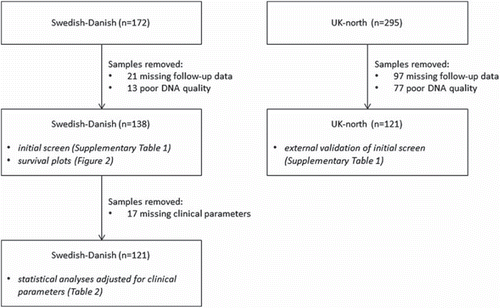
The cases studied were originally collected between September 2000 and February 2004 for three case-control studies that contributed to the INTERPHONE Study. Full details are provided in previously published literature [Citation3,Citation4]. In this study, the datasets are referred to as the Swedish-Danish dataset (172 glioblastoma cases) and the UK-north dataset (295 glioblastoma cases). Parts of these datasets have previously been included in studies that looked at both risk for glioma and outcome of glioblastoma [Citation5–8]. The Swedish-Danish dataset was supplemented with clinical data collected for surgery, chemotherapy, radiotherapy and survival. Survival data was also available for the UK-north dataset, but individual treatment data was not. The follow-up time for each patient was calculated as the time between date of diagnosis and date of death, or date of last follow-up in the medical record for living patients, or October 31, 2006, whichever occurred first. In the Swedish-Danish data, 21 cases were excluded from further analysis due to missing follow-up data, and an additional 13 cases were excluded due to poor DNA quality. In total, 138 cases were chosen for further analyses. Another 17 patients were missing values for at least one of the clinical parameters used for adjusting the statistical tests (i.e. extent of surgery, radiotherapy, chemotherapy, sex, country, and age at diagnosis). Hence these tests were performed on a subset consisting of 121 cases (). In the UK-north dataset, 97 cases were excluded from further analysis due to missing follow-up (all from the Trent region of northern UK) and an additional 77 cases were excluded due to poor DNA quality, leaving 121 cases for the final analyses (). For the Swedish-Danish dataset, the median follow-up was 12.9 months (range: 1.0–71.8 months); for the UK-north dataset, the median follow-up was 11.3 months (range: 2.2–78.8 months). See for descriptions and details of treatment.
Table I. Distribution of clinical characteristics.
Glioblastoma cases were identified through neurosurgery, neuropathology, oncology, and neurology centres and cancer registries. The following selection criteria were used: glioblastoma ICD 10th revision code 94403, aged 20 to 69 years in the Nordic countries or aged 18 to 69 years in the northern UK, and resident in the study region at time of diagnosis. Samples and clinicopathological information from participants were obtained after informed consent and with ethical review board approval in accordance with the tenets of the Declaration of Helsinki.
Selection of DNA repair genes and SNPs
The selection of SNPs has been described in detail elsewhere [Citation9]. Briefly, a list of over 35 000 candidate DNA repair polymorphisms was assembled through an inventory of 141 known DNA repair genes in current databases [Citation10–12]. Using an Illumina in house algorithm based on HapMap and LD select [Citation13,Citation14], we shortened the list to 2765 tag SNPs found in 136 genes (MAF > 0.1, r2 > 0.8 in HapMap CEPH (CEU) Utah residents with ancestry from northern and western Europe). After eliminating the SNPs unsuitable for the genotyping assay and further prioritising, we selected 1127 tag SNPs across 136 genes. This set of SNPs was supplemented with an additional 388 putative functional SNPs using the Illumina in house algorithm and PupaSuite web-based software (http://pupasuite.bioinfo.cipf.es/) [Citation15].
SNP genotyping
Standard methods were used to extract DNA from the collected blood samples, and DNA quantity was measured using the PicoGreen assay (Invitrogen Corp., Carlsbad, CA, USA). Protocols were standardised across all study centres. Next, the samples were genotyped by customised Illumina GoldenGate Arrays (Illumina Inc., San Diego, CA, USA). Samples with a corresponding GenCall score < 0.25 at any locus were set to “no call”. Genotyping of a sample was considered to have failed if it returned genotypes from less than 80% of all loci. Genotyping of an SNP was considered to have failed if it returned genotypes from less than 95% of all samples.
Statistical methods
In previous studies, genotypic frequencies in control subjects for each SNP were tested for departure from Hardy-Weinberg equilibrium (HWE) using an exact test [Citation9]. The hazard ratios for the heterozygous and rare homozygous variants were estimated in comparison with the common homozygous variant using Fisher's exact test in the Swedish-Danish dataset. Polymorphisms with a significant (p < 0.05) association to survival were selected for further validation in the UK-north dataset. The statistics for the SNPs displaying a significant (p < 0.05) association also in the UK-north dataset were subsequently adjusted for treatment – i.e. extent of surgery, radiotherapy, chemotherapy, sex, country and age at diagnosis – with a Cox proportional hazard ratio and a 95% confidence interval. The adjustment was performed in a subset of the Swedish-Danish dataset (). Polymorphisms found statistically significant (p < 0.05) after adjustment for treatment were considered final significant findings. Furthermore, p-values from the last test were adjusted by applying an empirical method based on permutations (n = 10 000), similar to the method described by Lystig [Citation16]. These adjustments were performed in R 2.11.1.
Results
This dataset, including information regarding genotyping success rates and quality, has been described in detail in a previous study by Bethke et al. [Citation9]. After eliminating SNPs that were not adequately genotyped and checking genotype failure rates as well as violations of the Hardy-Weinberg equilibrium, a total of 1458 SNPs remained for further analysis. This was one variable less than in Bethke's report [Citation9], because we had to remove one additional variable (rs4647404) due to genotyping failure among the samples in the Swedish-Danish dataset.
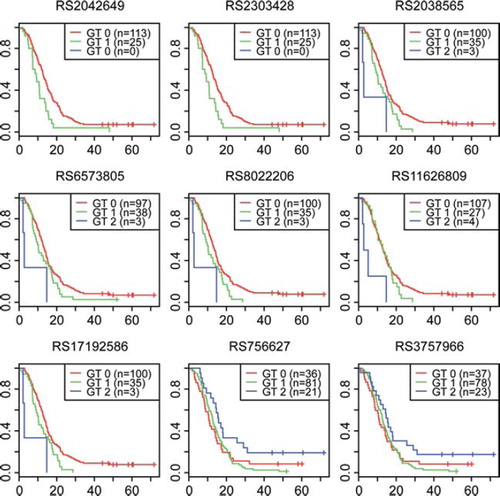
Out of the 1458 analysed SNPs, 142 were associated with outcome at the initial screening in the Swedish-Danish dataset as determined by Fisher's exact test at the 95% significance level (Supplementary Table I, http://informahealthcare.com/abs/doi/10.3109/0284186X.2011.616284). Validation analyses were conducted using an independent dataset (UK-north). Fifteen polymorphisms were found significantly associated with survival when subjected to external validation in the UK-north dataset with corresponding directions of increasing/decreasing odds ratios in both datasets. Subsequent adjustment of these 15 polymorphisms for country, sex, age, surgery (radical or not), radio- and chemotherapy using Cox regression in a subset of the Swedish-Danish dataset revealed nine SNPs with significant association to survival ( and Supplementary Table I). The subset consisted of the 121 patients where this complete set of information was available (). Two of the nine SNPs displayed significant p-values also following adjustment by permutation (). illustrates the survival discrepancies between the different gene variants; their respective median survival times are listed in . For the MSH2 SNP rs2042649, the median survival for patients who were homozygous for the common allele was 13.3 months, compared to 10.6 for patients with the heterozygous variant. The median survival for the RECQL4 rs756627 common allele was 10.6 months, compared to 15.8 for the rare allele.
Table II. Allelic variants associated with survival, based on 121 patients in the Swedish-Danish dataset with complete data.
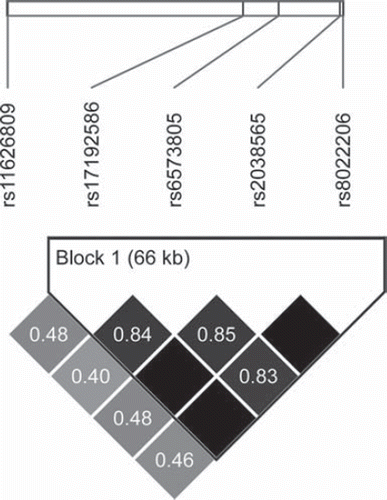
Five SNPs were found within the RAD51L1 gene (), all in strong LD with each other. Four of these – rs17192586, rs6573805, rs2038565, and rs8022206 – were located within the same haplotype block as calculated by the confidence interval algorithm in the Haploview 4.2 software [Citation17] (). On the observational level, they consequently shared an almost identical distribution with the same 32 individuals displaying a heterozygous genotype (an additional three patients were heterozygous for rs6573805), and the same three individuals displaying the rare homozygous genotype. The distribution was somewhat different for the fifth SNP in RAD51L1 (rs11626809), although the same three individuals were also homozygous for the rare allele.
Figure 3. Linkage disequilibrium (LD) plot of the five SNPs found significantly associated with survival, mapping to the RAD51L1 gene. The shading of each box represents r2 values, ranging from white (r2 = 0) to black (r2 = 1). The numeric r2 value is given within each box.
In genes RECQL4 and MSH2, two separate SNPs were found to be significantly associated with outcome. The two SNPs in each gene were in LD with each other (r2 = 1) in all instances, and accordingly the observational distribution was almost identical between the SNPs within the same gene.
Discussion
We have comprehensively studied a set of 1127 tag SNPs as well as 388 putative functional SNPs in DNA-repair genes to elucidate the potential association between genetic variation in these genes and glioblastoma outcome. Glioma patients’ response to standard chemo- and radiotherapy may largely be determined by the tumour's ability to repair DNA-damage. Thus knowledge regarding polymorphic variants in DNA-repair genes associated with outcome may influence future treatment decisions.
The nine SNPs that were found associated with survival were located in three different DNA-repair genes – RECQL4, MSH2, and RAD51L1. The SNPs within the same gene were generally in strong LD with each other and displayed identical or almost identical observational distribution. In most instances, a clear gene dosage effect could be observed, where patients who were homozygous for the rare allele displayed a significantly different hazard ratio compared to patients who were homozygous for the common allele (). These three genes are all implicated in different aspects of the intricate machinery that upholds genome integrity. Hence, one plausible explanation for our findings may be a differential response to DNA damaging therapy in glioblastoma patients with different genetic variants. Naturally, defects in these three genes – RECQL4, MSH2, and RAD51L1 – are typically linked to various cancers. RECQL4 belongs to the RecQ helicase family, which primarily functions to unwind double stranded DNA to allow for replication and DNA maintenance. RECQL4 is known to partake in DNA repair following UV-irradiation induced damage and is suggested to interact with RAD51, a key regulator of p53, to repair DNA double strand breaks [Citation18]. Mutations in this gene are known to be involved in the Rothmund-Thomson syndrome, characterised in part by elevated risk to develop osteosarcoma [Citation19]. MSH2 is part of the DNA mismatch repair mutS family. The protein initiates DNA repair through recognising and binding directly to mismatched nucleotides [Citation20]. MSH2 malfunction is known to be involved in both Lynch syndrome and Turcot syndrome, where some patients have an inherited predisposition for both glioma and colorectal cancer. Chemotherapy is shown to reduce the MSH2 protein expression levels in glioma [Citation21]. The RAD51 protein family, of which RAD51L1 is a member, is involved in repairing DNA double strand breaks through homologous recombination [Citation22]. It has been shown in animal studies that radioresistance is reduced by RAD51 inhibition [Citation2].
The SNPs reported to be significantly associated with survival in glioblastoma are for the most part mapped to gene introns, whereas non-synonymous SNPs included in the analysis did not show significant associations. For instance, of all the analysed SNPs within the RECQL4 gene, one was non-synonymous (rs2306386); however, it did not turn out to be significantly associated with survival in this study, whereas two polymorphisms in introns did (). Although SNPs mapped to introns most likely will not yield altered amino acid sequences, they may still alter gene regulation in terms of transcription, splicing, and transcript turnover rates. It is also possible that the polymorphisms presented here are not directly responsible for the observed effect on survival, but rather are in LD with causative gene variants.
Today information is limited regarding SNPs in the genes where we found potential correlations to cancer survival. Although previous studies have linked various SNPs in these genes to different cancers, the focus has largely been to find associations to cancer susceptibility. Recently, a genome-wide association study implicated a specific SNP in RAD51L1 for the risk of developing breast cancer [Citation23]. Although allelic variants of the genes described in our study have not been found to be related to an increased propensity to develop glioma, studies show that there are a number of low penetrance genes with such properties [Citation6,Citation8,Citation24], including genes in DNA repair pathways [Citation9]. With respect to survival, certain SNPs in each of RecQ1 and RAD54L – members of the same families as RECQL4 and RAD51L1, respectively – may be linked to outcome in pancreatic cancer [Citation25]. A few polymorphisms in other genes have been linked to survival in malignant glioma, e.g. gene variants of CX3CR1 [Citation26], as well as specific SNPs in the genes for IL4R and epidermal growth factor [Citation7,Citation27]. Moreover, minisatellites in the vicinity of the hTERT gene may be related to glioblastoma outcome, although no conclusive results exist [Citation5,Citation28]. Few studies have been confirmed in independent datasets. To our knowledge, however, genotype variants of the three genes described here have not previously been associated with glioblastoma survival.
In this study, all tissue samples were subject for pathology review before inclusion in the analysis. We believe this step to be necessary in order to avoid the risk of including other diagnoses and thereby possibly reaching erroneous conclusions. To further minimise the risk of erroneous conclusions, we adjusted our initial findings in an external test set to reduce the risk of false positive findings (type 1 errors). The concerns regarding false positive findings are particularly relevant for the allelic variants in RAD51L1 (rs2038565, rs6573805, rs11626809, rs17192586, and rs8022206) that were particularly rare, where only three or four patients displayed homozygosity for the rare genotype. The risk of spurious associations is naturally elevated when analysing rare alleles. This is an important factor to consider when evaluating our findings. In addition, we adjusted all p-values based on 10 000 permutations, which rendered only two significant variants (rs756627 and rs3757966) (). This, however, is likely to be a too stringent test, generating much the same results as Bonferroni correction. Relying entirely on this could result in many false negatives (type 2 errors). Therefore, we present both adjusted and unadjusted p-values.
In conclusion, we have revealed associations between SNPs in four separate genes within different DNA repair pathways and glioblastoma survival. Considering the importance of DNA-repair genes in the resistance to cancer treatment, we believe it is plausible that they are of functional importance and may constitute future drug targets, especially when it comes to enhancing the efficacy of radiotherapy.
http://informahealthcare.com/abs/doi/10.3109/0284186X.2011.616284
Download PDF (127.3 KB)Acknowledgements
The Northern UK Study thanks all study interviewers, administrators, computer programmers and the study Steering Group chaired by Professor David Coggon, along with the many neuropathologists, neuroradiologists, neurosurgeons, neurooncologists, clinical oncologists, neurologists, nurses and administrators in Scotland, West Midlands and West Yorkshire. The Swedish centre was supported by the Swedish Research Council, the Cancer Foundation of Northern Sweden, the Swedish Cancer Society, and the Nordic Cancer Union and the Umeå University Hospital Excellence grant. The Danish centre was supported by the Danish Cancer Society. Beatrice Melin was supported from Acta Oncologica foundation through the Royal Swedish Academy of Science. The cases were identified through the INTERPHONE study and supported by the European Commission Fifth Framework Program “Quality of life and Management of Living Resources” (contract number QLK4-CT-1999-01563) and the International Union against Cancer (UICC). The UICC received funds for this purpose from the Mobile Manufacturers’ Forum and GSM Association. Provision of funds to the INTERPHONE study investigators via the UICC was governed by agreements that guaranteed INTERPHONE's complete scientific independence. These agreements are publicly available at http://www.iarc.fr/ENG/Units/RCAd.html. The Northern UK Study centre received funding from the Mobile Telecommunications and Health Programme, the Health and Safety Executive, the Department of Health, the Scottish Executive, and the United Kingdom Network Operators (‘‘O2’’, Orange, T-Mobile, Vodafone, ‘‘3’’). The authors have no conflict of interest to declare.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Jeggo P, Lavin MF. Cellular radiosensitivity: How much better do we understand it? Int J Radiat Biol 2009;85: 1061–81.
- Ohnishi T, Taki T, Hiraga S, Arita N, Morita T. In vitro and in vivo potentiation of radiosensitivity of malignant gliomas by antisense inhibition of the RAD51 gene. Biochem Biophys Res Comm 1998;245:319–24.
- Cardis E, Richardson L, Deltour I, Armstrong B, Feychting M, Johansen C, . The INTERPHONE study: Design, epidemiological methods, and description of the study population. Eur J Epidemiol 2007;22:647–64.
- Hepworth SJ, Schoemaker MJ, Muir KR, Swerdlow AJ, van Tongeren MJ, McKinney PA. Mobile phone use and risk of glioma in adults: Case-control study. BMJ 2006;332: 883–7.
- Andersson U, Osterman P, Sjostrom S, Johansen C, Henriksson R, Brannstrom T, . MNS16A minisatellite genotypes in relation to risk of glioma and meningioma and to glioblastoma outcome. Int J Cancer 2009;125:968–72.
- Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, . Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet 2009;41:899–904.
- Sjöström S, Andersson U, Liu Y, Brönnström T, Broholm H, Johansen C, . Genetic variations in EGF and EGFR and glioblastoma outcome. Neuro-oncology Epub 2010 Mar 2.
- Andersson U, Schwartzbaum J, Wiklund F, Sjostrom S, Liu Y, Tsavachidis S, . A comprehensive study of the association between the EGFR and ERBB2 genes and glioma risk. Acta Oncol Epub 2010 May 7.
- Bethke L, Webb E, Murray A, Schoemaker M, Johansen C, Christensen HC, . Comprehensive analysis of the role of DNA repair gene polymorphisms on risk of glioma. Hum Mol Genet 2008;17:800–5.
- Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, . The UCSC Genome Browser Database. Nucleic Acids Res 2003;31:51–4.
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, . The human genome browser at UCSC. Genome Res 2002;12:996–1006.
- Wood RD, Mitchell M, Lindahl T. Human DNA repair genes, 2005. Mutat Res 2005;577:275–83.
- Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet 2004;74: 106–20.
- The International HapMap Project. Nature 2003;426: 789–96.
- Conde L, Vaquerizas JM, Dopazo H, Arbiza L, Reumers J, Rousseau F, . PupaSuite: Finding functional single nucleotide polymorphisms for large-scale genotyping purposes. Nucleic Acids Res 2006;34(Web Server issue):W621–5.
- Lystig TC. Adjusted P values for genome-wide scans. Genetics 2003;164:1683–7.
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, . The structure of haplotype blocks in the human genome. Science 2002;296:2225–9.
- Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J Cell Sci 2005;118(Pt 18):4261–9.
- Kitao S, Lindor NM, Shiratori M, Furuichi Y, Shimamoto A. Rothmund-thomson syndrome responsible gene, RECQL4: Genomic structure and products. Genomics 1999;61:268–76.
- Fishel R, Ewel A, Lescoe MK. Purified human MSH2 protein binds to DNA containing mismatched nucleotides. Cancer Res 1994;54:5539–42.
- Stark AM, Witzel P, Strege RJ, Hugo HH, Mehdorn HM. p53, mdm2, EGFR, and msh2 expression in paired initial and recurrent glioblastoma multiforme. J Neurol Neurosurg Psychiatry 2003;74:779–83.
- Kawabata M, Kawabata T, Nishibori M. Role of recA/RAD51 family proteins in mammals. Acta Medica Okayama 2005;59:1–9.
- Thomas G, Jacobs KB, Kraft P, Yeager M, Wacholder S, Cox DG, . A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet 2009;41: 579–84.
- Wrensch M, Jenkins RB, Chang JS, Yeh RF, Xiao Y, Decker PA, . Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet 2009;41:905–8.
- Li D, Frazier M, Evans DB, Hess KR, Crane CH, Jiao L, . Single nucleotide polymorphisms of RecQ1, RAD54L, and ATM genes are associated with reduced survival of pancreatic cancer. J Clin Oncol 2006;24:1720–8.
- Rodero M, Marie Y, Coudert M, Blondet E, Mokhrari K, Rousseau A, . Polymorphism in the microglial cell-mobilizing CX3CR1 gene is associated with survival in patients with glioblastoma. J Clin Oncol 2008;26:5957–64.
- Scheurer ME, Amirian E, Cao Y, Gilbert MR, Aldape KD, Kornguth DG, . Polymorphisms in the interleukin-4 receptor gene are associated with better survival in patients with glioblastoma. Clin Cancer Res 2008;14:6640–6.
- Carpentier C, Lejeune J, Gros F, Everhard S, Marie Y, Kaloshi G, . Association of telomerase gene hTERT polymorphism and malignant gliomas. J Neuro-oncol 2007;84:249–53.