Abstract
Background. Combined positron emission tomography (PET)/magnetic resonance imaging (MRI) is highly promising for biologically individualized radiotherapy (RT). Hence, the purpose of this work was to develop an accurate and robust registration strategy to integrate combined PET/MR data into RT treatment planning. Material and methods. Eight patient datasets consisting of an FDG PET/computed tomography (CT) and a subsequently acquired PET/MR of the head and neck (HN) region were available. Registration strategies were developed based on CT and MR data only, whereas the PET components were fused with the resulting deformation field. Following a rigid registration, deformable registration was performed with a transform parametrized by B-splines. Three different optimization metrics were investigated: global mutual information (GMI), GMI combined with a bending energy penalty (BEP) for regularization (GMI+ BEP) and localized mutual information with BEP (LMI+ BEP). Different quantitative registration quality measures were developed, including volumetric overlap and mean distance measures for structures segmented on CT and MR as well as anatomical landmark distances. Moreover, the local registration quality in the tumor region was assessed by the normalized cross correlation (NCC) of the two PET datasets. Results. LMI+ BEP yielded the most robust and accurate registration results. For GMI, GMI+ BEP and LMI+ BEP, mean landmark distances (standard deviations) were 23.9 mm (15.5 mm), 4.8 mm (4.0 mm) and 3.0 mm (1.0 mm), and mean NCC values (standard deviations) were 0.29 (0.29), 0.84 (0.14) and 0.88 (0.06), respectively. Conclusion. Accurate and robust multimodal deformable image registration of CT and MR in the HN region can be performed using a B-spline parametrized transform and LMI+ BEP as optimization metric. With this strategy, biologically individualized RT based on combined PET/MRI in terms of dose painting is possible.
Recently, combined imaging of positron emission tomography (PET) and magnetic resonance imaging (MRI) has become available [Citation1]. This new imaging technique allows for the simultaneous acquisition of functional MR and molecular PET information, resulting in accurately co-registered datasets. As a consequence, integrated PET/MR may be highly beneficial for radiotherapy (RT) individualization [Citation2]. On the one hand, the molecular, functional and anatomical information from PET/MR data be can be used in RT treatment planning (RTP) for improving the precision of tumor volume delineation [Citation3,Citation4]. Moreover, the molecular information on tumor physiology, biology, and radioresistance may serve as a basis for biologically individualized RT in terms of dose painting (DP) [Citation5,Citation6]. Furthermore, combined PET/MR imaging might be also valuable for the assessment of treatment response and follow-up after therapy [Citation7].
However, for the integration into RTP, PET/MR data has to be fused with the computed tomography (CT) image generally used for treatment planning. For an accurate fusion of PET/MR and CT data, the anatomical information of CT and MR can be used for registration, and the PET component of the PET/MR can be co-transformed with the resulting deformation field. Due to the different physical principles of image acquisition, CT and MR do not show a simple relationship between image intensities. Moreover, to date it is not possible to acquire combined PET/MR in the head and neck (HN) region with dedicated RT positioning aids. Therefore a suitable multimodal deformable registration (DR) algorithm is required. Another potential application of DR of MR and CT is the attenuation correction of PET/MR data using deformed CT images [Citation8].
Previous strategies for the DR of CT and MR images have been published for different anatomical sites such as liver [Citation9], breast [Citation10] and prostate [Citation11]. For the HN region, the development of DR strategies has so far mainly been focused on monomodal DR of CT images [Citation12]. Nevertheless, some studies also investigated DR algorithms for the fusion of multimodal imaging data. In the study published by Söhn et al., a multimodal DR algorithm based on rigidly matching local image subvolumes was applied to fuse CT and MR data of the HN region [Citation13]. Nevertheless, this study was intended as a proof of concept without quantitative evaluation. Another study applied a different algorithm matching the bony structures in combination with a linear elastic biomechanical finite element model for image registration of CT and MR in the HN region [Citation14], and a landmark-based evaluation for four datasets was provided.
A common approach for multimodal DR is to use an algorithm consisting of a B-spline parametrized transform and mutual information (MI) as similarity measure [Citation15,Citation16]. However, also localized versions of MI have been considered recently, where MI is evaluated in subregions of the images only [Citation17,Citation18].
In this study, B-spline-based algorithms with both global and localized forms of MI are applied for DR of CT and MR in the HN region and the registration accuracy is assessed by means of quantitative measures. The aim is to develop an accurate and robust registration method for potential future integration of PET/MR image data into RTP.
Material and methods
Patient data
Eight patient datasets acquired within a clinical imaging study were available. Each dataset consists of a PET/CT and subsequently acquired PET/MR, with the PET-tracer [18F]-Fluorodeoxyglucose (FDG) (cf. Supplementary Figure 1, to be found online at http://informahealthcare.com/doi/abs/10.3109/0284186X.2013.813964). The mean injected activity of FDG was 351 MBq (range 320–388 MBq). PET/CT data were acquired after a mean time of 81 min p.i. (range 76–94 min) and PET/MR data after 136 min p.i. (range 120–166 min). Detailed information about the patient characteristics is given in Supplementary Table I (to be found online at http://informahealthcare.com/doi/abs/10.3109/0284186X.2013.813964).
Figure 1. Fusion images after RR and DR for Patient 4. Fusion of the original CT (gray) and the deformed MR (orange) after RR (A) and after DR with LMI+ BEP (B). Fusion of the PET of the PET/CT (red) and the deformed PET of the PET/MR (green) after RR (C) and after DR with LMI+ BEP (D).
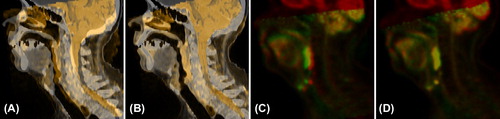
Table I. Quantitative results of the registration methods as mean (standard deviation) over all patients.
The PET/CT was acquired without fixation with a Siemens Biograph mCT (Siemens Healthcare, Knoxville, TN, USA). The CT consisted of a low-dose and contrast-enhanced acquisition. For PET reconstruction, three-dimensional (3D) OSEM with two iterations, 21 subsets and a 3D Gaussian filter of 2 mm was used. Approximate voxel sizes of the CT and the PET images were 0.8 × 0.8 × 3.0 mm3 and 1.6 × 1.6 × 3.0 mm3, respectively.
The PET/MR was acquired without fixation with the Siemens Biograph mMR (Siemens Healthcare, Erlangen, Germany). The MRI datasets taken into account for this study were T2-weighted acquisitions using a short time inversion recovery (STIR) sequence taken in the transversal direction. For PET reconstruction, 3D OSEM with three iterations, 21 subsets and a 3D Gaussian filter of 3 mm was used. MR attenuation correction was performed based on a segmentation approach on basis of spoiled gradient-echo sequences with DIXON-based fat-water separation. Approximate voxel sizes of the STIR and the PET images were 0.7 × 0.7 × 4.8 mm3 and 2.8 × 2.8 × 2.0 mm3, respectively.
Image registration
Image registration was performed using information from CT and MR only, with the CT serving as fixed image and the MR serving as moving image. After registration, both MR and PET from the PET/MR examination were transformed to the PET/CT data. Thus, after registration, the CT and the MR as well as the two PET images were defined in the same coordinate system.
First, a rigid registration (RR) was performed. The resulting transform was then used for the initialization of the DR methods, where three different strategies were investigated. Since the available datasets had different fields of view (FOV), they were cropped after RR in order to cover the same anatomical region.
Both RR and DR were performed with the freely available, open source registration package elastix [Citation19] which is based on the ITK (Insight Segmentation and Registration Toolkit, www.itk.org).
DR was performed with a three level multi resolution approach, using only Gaussian smoothing without downsampling. Smoothing scales were chosen as σ = 8.0,4.0,1.0 in x and y direction and σ = 2.0,1.0,0.5 in z direction, to account for voxel anisotropy.
For each DR strategy a transform parametrized by B-splines was used [Citation15]. For the optimization of the metric, a stochastic gradient descent optimizer [Citation20] was applied. In this optimization strategy, a sampler randomly chooses a given number of image positions every iteration for metric estimation
As similarity measure, firstly MI calculated from samples over the whole image domain was used, which is referred to as global MI (GMI). Secondly, a localized version of MI was considered, where the localization is achieved as described in [Citation17]. Briefly, sampling is constrained to a cubic subregion of the image of length Lsub, which is chosen randomly in every iteration step from the fixed image domain. This method is referred to as localized MI (LMI).
For regularization of the transform a bending energy penalty (BEP) term was applied [Citation15], defined as:
where are the voxel positions, and P is the number of voxels. Ti denotes the ith component of the transform, and x1, x2 and x3 are the coordinates in x, y and z direction, respectively. The BEP favors a smooth deformation field.
The following combinations were investigated for the registration metric M:
where λ determines the weight of the regularization with respect to the similarity measure.
For some of the free parameters of the DR methods, such as the B-spline grid spacing, the number of iterations and the number of samples used for metric evaluation during optimization, predetermined parameters were used. The remaining free parameters, i.e. the number of histogram bins used for MI calculation, λ, and Lsub, were optimized independently for each DR method according to the registration quality measures derived from segmentations described below. The final parameter sets are provided in Supplementary Table II (to be found online at http://informahealthcare.com/doi/abs/10.3109/0284186X.2013.813964).
Visual assessment of registration quality
A visual assessment of registration quality was performed based on fusion images of CT and MR as well as of the two PET images. Moreover, the deformation fields obtained from the different DR methods were assessed qualitatively by the corresponding maps of Jacobian determinants IJac [Citation21,Citation22]. Jacobian determinant values smaller than 1 are associated with a local volume compression, values equal to 1 with local volume preservation, and values greater than one with local volume expansion. Negative values of the Jacobian determinant are obtained in the presence of foldings, which are unrealistic for medical images.
Quantitative assessment of registration accuracy
Several measures for the determination of registration accuracy were implemented using ITK and VTK (Visualization Toolkit, www.vtk.org). For the structures skin, carotids, and respiratory tract segmented on CT and MR, both the Dice similarity index (DSI) and mean distances after registration were evaluated. As mean distance measures, the mean volume distance (MVD) was determined for the skin as well as for the respiratory tract structure, whereas for the carotids the mean line distance (MLD) was evaluated. In addition, the non-overlapping fraction (NOF) of the bony structures segmented from CT and the spinal canal segmented from the MR image was calculated. Moreover, the mean residual distance [mean point distance (MPD)] of anatomical landmarks defined by two experienced radiation oncologists was evaluated after registration.
A validation of the local registration accuracy in the tumor region based on the image information from CT and MR is difficult, since the intra-tumor region can be of low contrast in these imaging modalities. As the PET images provide local image information in this region, the registration accuracy in the tumor region was assessed by normalized cross correlation (NCC) of the fused PET images.
Detailed information about the quantitative registration accuracy measures is provided in the supplementary material (to be found online at http://informahealthcare.com/doi/abs/10.3109/0284186X.2013.813964).
Results
Qualitative evaluation
Fusion images of CT and MR as well as of the PET images are displayed in exemplarily for Patient 4 after both RR and DR with LMI+ BEP. The fusion images after RR showed large remaining misalignments due to different patient positioning. After DR, a visually good alignment of both the anatomical and the functional images could be obtained.
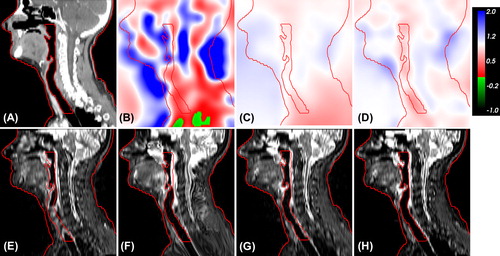
shows IJac and the corresponding deformed MR images obtained by the three DR methods for Patient 1. GMI led to Jacobian determinant values with high absolute differences to 1, and also foldings were present. Compared to RR, the corresponding deformed MR was better aligned to the shown anatomical contours, but unrealistic deformations occurred. The deformed MR images obtained by the registration strategies that apply a regularization term (GMI+ BEP and LMI+ BEP) did not suffer from unrealistic deformations and corresponding IJac showed only moderate and smooth volume expansions and compressions. Moreover, also a good alignment to the anatomical contours was achieved.
Figure 2. Map of Jacobian determinants (IJac) and corresponding deformed MR from different registration methods for Patient 1. Original CT (A), IJac from DR with GMI (B), GMI+ BEP (C) and LMI+ BEP (D). Transformed MR from RR (E), deformed MR from DR with GMI (F), GMI+ BEP (G) and LMI+ BEP (H). The structures skin and respiratory tract segmented on the original CT are shown as red contours.
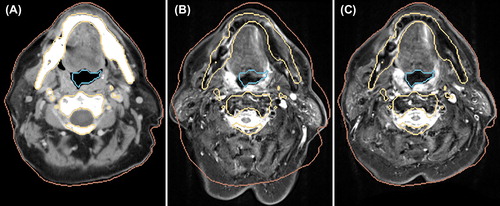
shows axial slices of the original CT and the transformed MR after RR as well as after DR with LMI+ BEP for Patient 2, in addition to contours defined on basis of the CT image. For the rigidly transformed MR, large misalignments to the CT contours remained, whereas the DR with LMI+ BEP led to a significant improvement. However, for the skin some misalignments remained in regions where large local deformations are present.
Quantitative evaluation
Quantitative results for all patients are summarized in as well as in . For GMI, the measures for the skin improved compared to RR, but unrealistic deformations as shown in led to worse quantitative registration results for other measures. For GMI+ BEP, all registration accuracy measures improved compared to RR. However, considerable variations remained, showing that the method is not robust with respect to inter-patient variation between different datasets.
Figure 4. Quantitative results of the registration methods. Left: Boxplots of quality measures ranging between 0 and 1, with 1 being the best value. Right: Boxplots of distance quality measures, with values given in mm. Low values indicate good registration accuracy. Outliers are shown as black crosses.
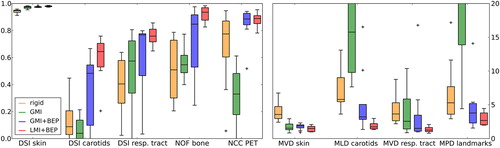
In contrast, LMI+ BEP showed a robust behavior and good registration accuracy. Residual distances were in the order of half the voxel size of fixed and moving image in z-direction (voxel size CT: 3.0 mm, MR: 4.8 mm) (cf. ). Moreover, the NCC of the PET images in the tumor region improved in line with the registration accuracy measures defined on CT and MR.
Discussion
In this work, three different DR strategies for the fusion of CT and MR in the HN region were evaluated qualitatively and quantitatively, with the purpose of integrating PET/MR data into RTP. All algorithms used a multi-resolution approach and a B-spline parametrized transform, whereas the different metrics GMI, GMI+ BEP and LMI+ BEP were applied. Compared to RR, using the metric GMI in most cases resulted in worse registration results in addition to unrealistic deformations. In principle, the degree of unrealistic deformations could be lowered by a smaller B-spline grid spacing, but this would simultaneously decrease the degree of freedom of the transform. For DR with GMI+ BEP, registration accuracy improved compared to RR, but a lack of robustness was observed. Using LMI+ BEP as optimization metric, accurate and robust results were obtained, even for patients with large positioning differences in CT and MR. Distance quality measures showed that residual distances were in the order of half the voxel size of the CT and MR in z-direction, indicating a high geometric accuracy.
Due to the small diameter of the carotids, the DSI for this structure can decrease severely even for small misregistrations. Therefore, the MLD should provide a better measure of registration accuracy for this structure, showing that the registration accuracy in the carotid region is similar to the other structures. In contrast to the other registration methods, for LMI+ BEP no outliers were present apart from the outlier observed for the DSI measure of the carotids (cf. ). However, since this registration method has been evaluated on a limited dataset, at least a visual examination of the registration results obtained with LMI+ BEP should be performed for additional datasets.
However, there are also inherent limitations of the algorithm as shown in . Since the B-spline parametrization provides only a limited degree of freedom of the transform, the algorithm is not able to map large local deformations. This ability is further reduced by the BEP. However, both the parametrization and the BEP favor a smooth transform which generally is a reasonable assumption for medical images, particularly in low-contrast regions where little anatomical information is available [Citation24].
Especially in the case of MRI data, spatial intensity distortions may be present. Hence, using the localized instead of the global form of MI is preferential in this case as it evaluates the MI in subregions of the images only [Citation18]. LMI is advantageous also for multimodal registration if one intensity class corresponds to a specific tissue type in one imaging modality and to different tissue types in the other imaging modality [Citation18]. Finally, if only a limited number of samples are chosen during optimization to evaluate MI as performed in this study, choosing the samples from a localized region may improve the statistical power of the method.
For clinical application, high registration accuracy in the tumor region is of major importance. Since there was a temporal delay between the acquisitions of the two PET images in our study, it is not expected that voxels of the same anatomical position have the same intensity values, but the assessment of the correlation of the PET images after registration still provides a meaningful measure of correspondence. The high NCC between the PET images indicates that surrounding anatomical structures in CT and MR give sufficient information for the registration in the low contrast tumor region.
In principle, the proposed registration method could also be applied to other anatomical sites. However, a re-optimization of the free parameters of the registration method may be favorable in this case to meet the different conditions of the specific body region.
PET/MR data can be integrated into biologically adapted RT in different ways. Using contours derived from PET and MR [Citation25] requires high registration accuracy around the contour boundaries only. The situation is different if data is to be integrated on a voxel basis as for dose paining by numbers (DPBN) [Citation6]. In that case, intra-tumor registration accuracy is of the highest importance.
To date, it is not possible to acquire simultaneous PET/MR data in the HN region with RT positioning aids. Therefore, it is beneficial that LMI+ BEP yields robust results also in the case of larger positioning differences between CT and MR. Nevertheless, dedicated RT positioning aids compatible with PET/MR would be favorable, since the final geometric accuracy achieved with DR will be improved by a better initial alignment. By using appropriate positioning aids also strong local deformations could be avoided, which cannot be correctly mapped by the proposed algorithm due to its design. However, even with positioning aids it is likely that slight deformations remain, and a DR in addition to a RR may still be favorable. Particularly for the evaluation of treatment response, DR allows to account for shrinkage of the tumor. For final integration of PET/MR data into RTP, the definition of PET as well as MR acquisition parameters should be optimized to meet special RT requirements, such as a high, isotropic resolution and correspondingly adjusted voxel sizes.
In conclusion, this study showed that DR with a B-spline parametrized transform combined with LMI+ BEP as optimization metric yields accurate and robust results for registration of CT and MR in the HN region. As a consequence, this strategy for deformable multimodal image registration provides a basis for the integration of individual molecular, functional and anatomical PET/MR data into RTP.
Supplementary Tables I and II
Download PDF (3.9 MB)Acknowledgements
This work was supported by the German Research Foundation, grant no. AL 877/1–3 and by intramural funding of the University Hospital Tübingen, fortüne grant no. 1945-0-0. DT was financially supported by the European Social Fund and the Ministry of Science, Education and the Arts Baden-Württemberg.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Pichler BJ, Kolb A, Nägele T, Schlemmer HP. PET/MRI: paving the way for the next generation of clinical multimodality imaging applications. J Nucl Med 2010;51:333–6.
- Thorwarth D, Leibfarth S, Mönnich D. Potential role of PET/MRI in radiotherapy treatment planning. Clin Transl Imaging 2013;1:45–51.
- Shepherd T, Teras M, Beichel RR, Boellaard R, Bruynooghe M, Dicken V, et al. Comparative study with new accuracy metrics for target volume contouring in PET image guided radiation therapy. IEEE Trans Med Imaging 2012;31:2006–24.
- Groenendaal G, Borren A, Moman MR, Monninkhof E, van Diest PJ, Philippens ME, et al. Pathologic validation of a model based on diffusion-weighted imaging and dynamic contrast-enhanced magnetic resonance imaging for tumor delineation in the prostate peripheral zone. Int J Radiat Oncol Biol Phys 2012;82:e537–44.
- van der Heide UA, Houweling AC, Groenendaal G, Beets-Tan RGH, Lambin P. Functional MRI for radiotherapy dose painting. Magn Reson Imaging 2012;30:1216–23.
- Thorwarth D, Geets X, Paiusco M. Physical radiotherapy treatment planning based on functional PET/CT data. Radiother Oncol 2010;96:317–24.
- Andersen EKF, Kristensen GB, Lyng H, Malinen E. Pharmacokinetic analysis and k-means clustering of DCEMR images for radiotherapy outcome prediction of advanced cervical cancers. Acta Oncol 2011;50:859–65.
- Hofmann M, Steinke F, Scheel V, Charpiat G, Farquhar J, Aschoff P, et al. MRI-based attenuation correction for PET/MRI: A novel approach combining pattern recognition and atlas registration. J Nucl Med 2008;49:1875–83.
- Osorio EMV, Hoogeman MS, Romero AM, Wielopolski P, Zolnay A, Heijmen BJM. Accurate CT/MR vessel-guided nonrigid registration of largely deformed livers. Med Phys 2012;39:2463–77.
- Dmitriev ID, Loo CE, Vogel WV, Pengel KE, Gilhuijs KGA. Fully automated deformable registration of breast DCE-MRI and PET/CT. Phys Med Biol 2013;58: 1221–33.
- Lian J, Xing L, Hunjan S, Dumoulin C, Levin J, Lo A, et al. Mapping of the prostate in endorectal coil-based MRI/MRSI and CT: A deformable registration and validation study. Med Phys 2004;31:3087–94.
- Castadot P, Lee JA, Parraga A, Geets X, Macq B, Grégoire V. Comparison of 12 deformable registration strategies in adaptive radiation therapy for the treatment of head and neck tumors. Radiother Oncol 2008;89:1–12.
- Söhn M, Birkner M, Chi Y, Wang J, Yan D, Berger B, et al. Model-independent, multimodality deformable image registration by local matching of anatomical features and minimization of elastic energy. Med Phys 2008;35:866–78.
- du Bois d’Aische A, De Craene M, Geets X, Grégoire V, Macq B, Warfield SK. Estimation of the deformations induced by articulated bodies: Registration of the spinal column. Biomed Signal Process Control 2007;2:16–24.
- Rueckert D, Sonoda L, Hayes C, Hill D, Leach M, Hawkes D. Nonrigid registration using free-form deformations: Application to breast MR images. IEEE Trans Med Imaging 1999;18:712–21.
- Mattes D, Haynor D, Vesselle H, Lewellen T, Eubank W. PET-CT image registration in the chest using free- form deformations. IEEE Trans Med Imaging 2003; 22:120–8.
- Klein S, van der Heide UA, Lips IM, van Vulpen M, Staring M, Pluim JPW. Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Med Phys 2008;35:1407–17.
- Loeckx D, Slagmolen P, Maes F, Vandermeulen D, Suetens P. Nonrigid image registration using conditional mutual information. IEEE Trans Med Imaging 2010;29:19–29.
- Klein S, Staring M, Murphy K, Viergever M, Pluim J. Elastix: A toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 2010;29:196–205.
- Klein S, Pluim JPW, Staring M, Viergever MA. Adaptive stochastic gradient descent optimisation for image registration. Int J Comput Vis 2009;81:227–39.
- Christensen G, Johnson HJ. Consistent image registration. IEEE Trans Med Imaging 2001;20:568–82.
- Fiorino C, Maggiulli E, Broggi S, Liberini S, Cattaneo GM, Dell’Oca I, et al. Introducing the jacobian-volume-histogram of deforming organs: Application to parotid shrinkage evaluation. Phys Med Biol 2011;56:3301–12.
- Lorensen WE, Cline HE. Marching cubes: A high resolution 3D surface construction algorithm. SIGGRAPH Comput Graph 1987;21:163–9.
- Fischer B, Modersitzki J. Ill-posed medicine—an introduction to image registration. Inverse Probl 2008;24:034008.
- Thorwarth D, Henke G, Müller AC, Reimold M, Beyer T, Boss A, et al. Simultaneous 68Ga-DOTATOC-PET/MRI for IMRT treatment planning for meningioma: First experience. Int J Radiat Oncol Biol Phys 2011;81:277–83.