Abstract
There is growing evidence that sunitinib plasma levels have an impact on treatment outcome in patients with metastatic renal cell carcinoma (mRCC). We studied the impact of single nucleotide polymorphisms (SNPs) in genes involved in sunitinib pharmacokinetics, and additionally, sunitinib pharmacodynamics on dose reductions of the tyrosine kinase inhibitor.
Methods. We retrospectively analyzed germ-line DNA retrieved from mRCC patients receiving sunitinib as first-line therapy. We genotyped 11 key SNPs, respectively, in ABCB1, NR1/2, NR1/3 and CYP3A5, involved in sunitinib pharmacokinetics as well as VEGFR1 and VEGFR3, which have been suggested as regulators of sunitinib pharmacodynamics. Association between these SNPs and time-to-dose-reduction (TTDR) was studied by Cox regression.
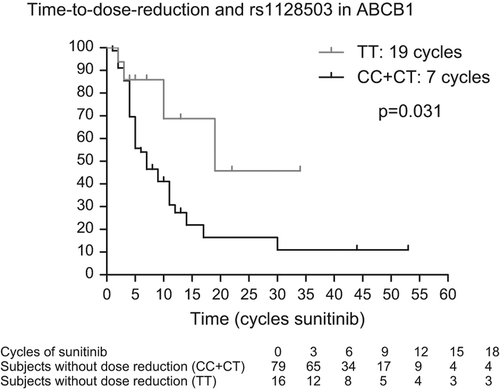
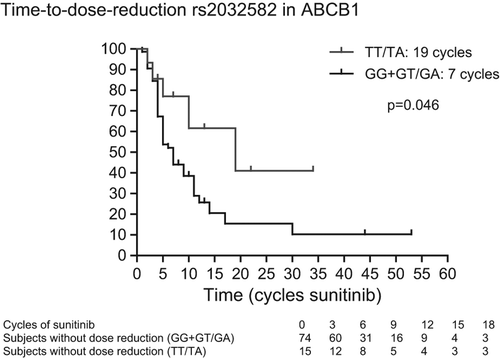
Results. We identified 96 patients who were treated with sunitinib and from whom germ-line DNA and data on dose reductions were available. We observed an increased TTDR in patients carrying the TT-genotype in ABCB1 rs1125803 compared to patients with CC- or CT-genotypes (19 vs. 7 cycles; p = 0.031 on univariate analysis and p = 0.012 on multivariate analysis) and an increased TTDR in patients carrying the TT/TA-variant in ABCB1 rs2032582 compared to patients with the GG- or GT/GA-variant (19 vs. 7 cycles; p = 0.046 on univariate analysis and p = 0.024 on multivariate analysis).
Conclusion. mRCC patients carrying the rs1128503 TT-variant or the TT/TA-variant in rs2032582 in ABCB1, which encodes for an efflux pump, do require less dose reductions due to adverse events compared to patients with the wild type or heterozygote variants in these genes.
Inactivation of the von Hippel-Lindau (VHL) tumor suppressor gene is the most frequent molecular alteration in clear cell renal cell carcinoma (RCC). Inactivated VHL leads to elevated protein levels of hypoxia-induced factor-α which upregulates the vascular endothelial growth factor (VEGF) and platelet-derived growth factor (PDGF) pro-angiogenic signaling pathways. Targeted therapies directed against the VEGF- and PDGF-receptor have significantly improved the outcome of patients with metastatic renal cell carcinoma (mRCC). Sunitinib malate is an orally administered tyrosine kinase receptor inhibitor (TKI) that targets VEGF and PDGF receptors, KIT, FLT-3, colony stimulating factor-1 receptor, and RET. In a randomized controlled trial sunitinib significantly prolonged progression-free survival (PFS) (11 vs. 5 months, p < 0.001) as compared to interferon alpha [Citation1]. Median overall survival (OS) was 26.4 and 21.8 months, respectively (p = 0.051) [Citation2]. Sunitinib is a current standard treatment option in mRCC, but other anti-VEGFR and anti-PDGFR-targeted TKIs like sorafenib, pazopanib and axitinib are also used in certain clinical settings.
Although 50% of RCC patients receiving sunitinib experience an objective response and 43% achieve disease stabilization, 7% will experience progressive disease (PD) at first evaluation probably due to intrinsic resistance or due to other factors [Citation2]. Moreover, even patients with an initial clinical benefit will finally progress due to acquired resistance or for other reasons. Although different mechanisms of primary and secondary resistance have been proposed, reliable biomarkers predictive of sunitinib sensitivity or primary/secondary resistance are still lacking [Citation3].
Sunitinib plasma levels are not dosed in clinical routine, even though it is known that sunitinib plasma levels might impact efficacy of sunitinib treatment in mRCC. A population pharmacokinetic analysis of sunitinib and its primary active metabolite, SU12662, found that the pharmacokinetics of both compounds were significantly influenced by several covariates including gender, age, and weight; however, the magnitude of the predicted changes in exposure minimized the necessity for dose adjustments [Citation4]. A meta-analysis of pharmacokinetic data from 443 patients treated with sunitinib showed that higher plasma levels of sunitinib and its active metabolite SU12662 were associated with prolonged time-to-tumor-progression (TTP) and OS [Citation5]. Other studies have shown that the occurrence of adverse events, in particular hypertension, is possibly linked to improved treatment outcome [Citation6]. Finally, several studies in RCC have shown associations between polymorphisms in genes linked to sunitinib pharmacokinetics and outcome on sunitinib [Citation7–10] or pazopanib [Citation11,Citation12].
The main objective of the present study was to analyze the impact of SNPs in selected genes potentially linked to sunitinib pharmacokinetics (ABCB1, NR1/2, NR1/3 and CYP3A5) and the occurrence of dose reductions during treatment. Additionally, we analyzed the impact of SNPs in two genes encoding sunitinib targets (VEGFR1 and VEGFR3), linked to sunitinib efficacy, and the occurrence of dose reductions.
Materials and methods
For the purpose of this retrospective study, germ-line DNA samples were collected from the “CIT-rein” kidney tumor bank and from patients treated at the University Hospitals Leuven. The French-Belgian multicentric “CIT-rein” kidney tumor bank contains more than 250 frozen pathologically confirmed RCC tumor samples collected at 20 academic hospitals. In the “CIT-rein” kidney tumor bank, we selected the samples of patients treated in first-line with sunitinib at a starting dose of 50 mg/day four weeks on, two weeks off and of whom frozen normal kidney tissue as well as data on dose reductions were available. In order to extend the series, we sampled peripheral blood in all the RCC patients treated at the University Hospitals Leuven from July 2011 to December 2012 applying the inclusion criteria. Eligible patients could have received cytokines as systemic treatment for kidney tumors, but they could not have received any other TKI or mammalian target of rapamycin (mTOR) inhibitor before starting sunitinib.
Dose reduction policy and timing of clinical radiological assessments were left to the discretion of the attending doctors in accordance with current local practice guidelines. Usually, whenever necessary for tolerance issues, in a first step, the dose is reduced to 37.5 mg/day and, if necessary, in a second step to 25 mg/day. In some patients, sunitinib is definitively stopped for tolerance issues.
The endpoint of this study was time-to-dose- reduction (TTDR), calculated as the time between the start of sunitinib and the occurrence of a dose reduction to 37.5 mg/day or of definitive stop of sunitinib for tolerance issues. Therefore, in this study, the SNPs were primarily evaluated as toxicity-related markers, although it was also foreseen to check our previous findings on associations between these SNPs and outcome in this patient series in order to show the inverse correlation between TTDR and outcome. If a patient's regimen was switched from 50 mg/day to 37.5 mg/day continuously because of flare-up during the two weeks off sunitinib, this was not considered as a dose reduction for adverse event and the censing was closed at the moment of dose adaptation.
For the statistical analysis, the genotypes were combined as much as possible, as it was done in the original publications. Details and exceptions to this rule are documented in the legend of .
For those SNPs that were significantly associated with TTDR, we also analyzed their association with PFS and OS. For this efficacy analysis, only patients with clear cell RCC were considered as previous publications on associations between polymorphisms and efficacy were only reported in clear cell RCCs. Moreover, for the efficacy analysis, all the patients had to complete at least one cycle of sunitinib and had to reach at least the first evaluation by computed tomography (CT) scan. Response evaluation was done by RECIST in most of the cases.
The protocol was approved by the medical ethics review boards of all participating institutions, and signed consent was obtained from all patients. In some cases, we used frozen biologic material from patients who had already died and for whom a general positive advice for the utilization of remaining tissue was foreseen by the institutional board.
SNPs with potential relevance for sunitinib dose reductions were selected from the literature ( and ). In particular, we included SNPs in genes linked to sunitinib or pazopanib pharmacokinetics or pharmacodynamics associated with efficacy and/or dose reductions in previous publications with sunitinib [Citation7–10] or pazopanib [Citation11,Citation12].
Table IA. Analyzed SNPs linked to sunitinib pharmacokinetics.
Table IB. Analyzed SNPs linked to sunitinib pharmacodynamics.
DNA was isolated from fresh frozen normal kidney tissue sampled in the nephrectomy specimen using the Qiaquick extraction kit (Qiagen, Valencia, CA, USA) and quantified by fluorometry (Fluoroskan Thermo Labsystems, Cergy-Pontoise, France). DNA was isolated from peripheral blood with the Qiagen DNA kit (Qiagen) and the final DNA concentration was quantified with Nanodrop (Nanodrop, Wilmington, DE, USA). High-throughput SNP genotyping was performed using the Sequenom MassArray platform [Citation13]. Investigators blinded for the clinical data performed the genotyping analysis. Overall, the 11 selected SNPs were successfully genotyped with success rates ≥ 92% for each SNP and an overall average success rate of 98%. For most of the SNPs, genotypes were analyzed in the same way as they were described in the original reports (i.e. according to dominant, recessive or co-dominant genetic models or in the context of a specific haplotype). Details are given in .
Clinical data were collected at 12 different sites in France (11) and Belgium (1). TTDR, PFS and OS were calculated by Cox regression. Based on the data of Houk et al. [Citation4], we considered that gender, age and general shape of the patient as reflected by his IMDC prognostic score could influence tolerance and as a consequence TTDR. These factors were tested in univariate analysis, except patient weight which was not available. Any parameter related to TTDR in univariate analysis by Kaplan-Meier with a p-value < 0.2 was included in the multivariate model (Cox regression). Without correction for multiple testing, results with a p-value of < 0.05 were considered as significant. However, correction for multiple testing by Bonferroni, taking into account the fact that the correlation with 11 SNPs was analyzed, indicated a p-value of < 0.005 as the threshold for significance. Statistical analyses were conducted using GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA) and XLSTAT software (Addinsoft, Paris, France).
Results
We used tissue and clinical information from 96 patients who started sunitinib between November 2005 and November 2012 and closed the follow-up database in June 2013. For 72 patients, frozen normal kidney samples from the “CIT-rein” kidney tumor bank were used and for 24 additional patients treated in Leuven, peripheral blood was used. The data of 81 of these patients were used in a previous publication on the impact of ABCB1 polymorphisms on outcome by our own group [Citation8]. shows the clinical characteristics of these patients. Mean age at diagnosis was 59 years (range 25–84). The majority of patients (> 95%) were of Caucasian origin. According to the International Metastatic Renal Cell Carcinoma Database Consortium (IMDC) prognostic criteria [Citation14], 14% of patients were categorized into the favorable risk group, 59% had intermediate and 27% poor risk.
Table II. Patient characteristics at diagnosis of mRCC and at the start of sunitinib treatment.
Forty-nine of 96 patients (51%) required dose reductions or definitive stop of sunitinib after a median TTDR of five cycles (46 dose reduction and 3 treatment withdrawal). Median TTDR in all patients, i.e. in those undergoing dose reduction and those not undergoing dose reduction, was nine cycles. The most frequent reason for dose reduction were hand foot skin reactions (17 patients), followed by diarrhea (14 patients), fatigue (9 patients), arterial hypertension (5 patients) and thrombocytopenia (5 patients). Less common reasons for dose reductions were anorexia, cardiotoxicity, mucositis, nausea and neutropenia. At the time of analysis, 71 (74%) patients had progressed and 59 (61%) had died. The median follow-up was 59 months (range 2–89 months) after the start of sunitinib. The median PFS of the whole study population was 15 months and the median OS 29 months. Best response could be evaluated in 91 patients. Seven of 91 (7.7%) patients had a complete response (CR), 33/91 (36.3%) patients a partial response (PR), 37/91 (40.7%) stable disease (SD) and 14/91 (15.4%) PD as best response.
For each of the 11 genotyped polymorphisms the respective genotypes, allele frequencies and changes at the amino acid level are described in . The observed allele frequencies for each polymorphism were similar as previously reported in the dbSNP database (dbSNP build 136) or 1000 Genomes Project, except for SNPs rs2276707.
Table III. Genotype and allele distribution of selected SNPs.
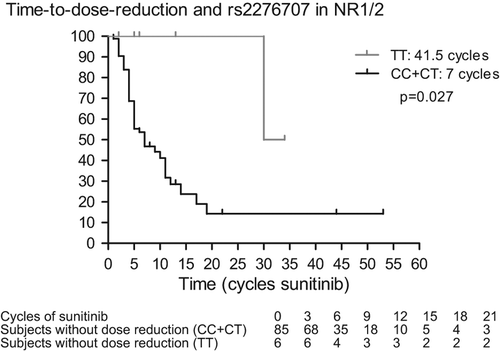
The association between these polymorphisms and TTDR, as assessed by univariate analysis, are reported in and displayed in . We observed increased TTDR in patients carrying the rs1125803 TT-genotype compared to patients carrying the CC- or CT-genotypes in ABCB1 (19 vs. 7 cycles; p = 0.031). Likewise, we observed increased TTDR in patients carrying the TT/TA-variant in ABCB1 rs2032582 (19 vs. 7 cycles; p = 0.046), but rs1128503 and rs2032582 were in high linkage disequilibrium (r² = 0.984) with each other. We also observed increased TTDR in patients with the TT-genotype in NR1/2 rs2776707 compared to patients with CC- and CT-genotypes (41.5 vs. 7 cycles; p = 0.027). We could not observe any association between SNPs in NR1/3, CYP3A5, VEGFR1 and VEGFR3 and TTDR.
Table IV. Univariate analysis: association between SNPs and time of dose reduction.
In view of an adjusted p-value and of the multivariate analysis, we checked TTDR in female and male (10 vs. 9 months; p = 0.13) and in IMDC good and intermediate versus poor risk patients (11 vs. 7 months; p = 0.054). Age at start of sunitinib (under or above the median age of 61 years) had no influence on TTDR [HR 0.89 (95% CI 0.49–1.60); p = 0.69]. Unfortunately, patient weight at start of sunitinib therapy was not available in a considerable part of the patients. Taking in to account gender and IMDC, the adjusted p-value is 0.014 for rs1128503 in ABCB1, 0.025 for rs2032582 in ABCB1 and 0.063 for rs2776707 in NR1/2.
In a next step, we introduced the other polymorphism in the multivariate analysis. Including gender, IMDC (good and intermediate vs. poor), rs1128503 in ABCB1 and rs2776707 in NR1/2, the p-value for the association between these SNPs and TTDR were 0.012 and 0.058, respectively. With rs2032582 (in ABCB1 instead of rs1128503) and rs2776707, the p-value for the association between these SNPs and TTDR were 0.024 and 0.060, respectively. Note that rs1128503 and rs2032582 were not included in the same multivariate analysis, because of their high linkage disequilibrium (r² = 0.984).
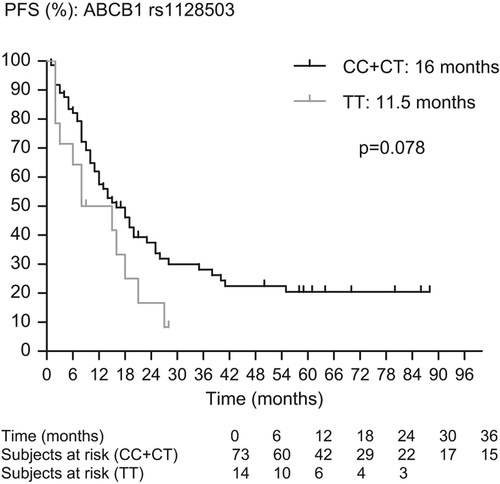
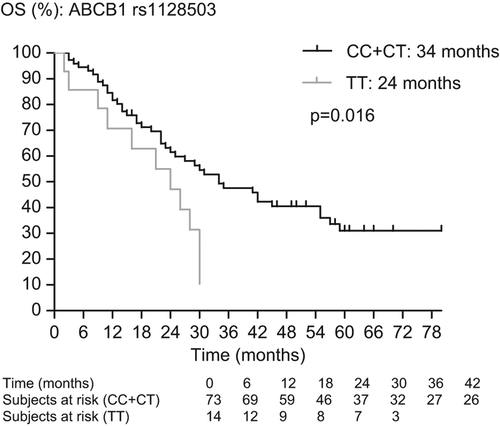
In a previous publication, we showed the impact of SNP rs1128503 in ABCB1 on outcome in mRCC treated with sunitinib. In order to show the inverse correlation between TTDR and outcome, we checked the impact of the SNPs associated with dose reductions on outcome. As there is no complete overlap with the previously published series, we report the outcome data of the present patient series. In patients with clear cell histology, we have found a trend to a shorter PFS (11.5 vs. 16 months, p = 0.078) and a shorter OS (24 vs. 34 months, p = 0.016) in patients with the TT-genotype compared to patients with the CC- and CT-genotypes in rs1125803 in ABCB1, a trend to a shorter PFS (15 vs. 18 months, p = 0.094) and a shorter OS (26 vs. 41 months, p = 0.012) in patients with the TT/TA-genotype compared to patients with the GG- and GA/GT-genotype in rs2032582 in ABCB1 and a shorter PFS (7 vs. 18 months; p = 0.011) and a trend to a shorter OS (12 vs. 31 months; p = 0.14) in patients with the TT-genotype compared to patients with the CC- and CT-genotypes in rs2776707 in NR1/2 ( and ).
Discussion
The main objective of the present study was to analyze the impact of SNPs in selected genes potentially linked to sunitinib pharmacokinetics (ABCB1, NR1/2, NR1/3 and CYP3A5) and the occurrence of dose reductions during treatment. We hypothesized that patients carrying genotypes that reduce absorption of sunitinib or increase metabolism of sunitinib – through lower sunitinib plasma levels and less frequent adverse events – less frequently require dose reductions.
In a series of 96 mRCC patients treated with sunitinib as first-line targeted therapy, we observed an association between SNP rs1128503 in ABCB1, rs2032582 in ABCB1 as well as SNP rs2776707 in NR1/2 and the time point of dose reductions during sunitinib treatment, although the latter was not confirmed on multivariate analysis. Our time-to-event approach enabled us to avoid lead-time bias, which could easily have occurred if we would have merely compared the incidence of dose reductions in subgroups with significantly different treatment durations. The impact of these SNPs on PFS and OS was also analyzed on the patients’ series, showing an inverse correlation between efficacy and dose reductions. Note that in a previous publication, we had already reported on the association between these SNPs and outcome on a patient series including 81 patients of the present study. With the lack of a placebo-treated control group, we cannot define if these SNPs have a prognostic or a predictive value for outcome, although the fact that these genes are involved in sunitinib pharmacokinetics points toward a predictive value.
At the start of therapy, anti-VEGFR-TKIs are generally administered at a fixed dose irrespective of the age, gender, weight or length of the patient. In the case of sunitinib, the starting dose is 50 mg/day for four weeks, followed by two weeks off-treatment. Many patients require dose modifications, i.e. dose reductions to 37.5 mg/day or even 25 mg/day due to tolerance issues. In the pivotal sunitinib trial, 38% of patients had dose interruptions and 32% had dose adaptations due to toxicity [Citation1]. Remarkably, Houk et al. observed that when doses are lowered to 37.5 mg/day or subsequently even to 25 mg/day due to tolerance issues, or even when the dose of sunitinib is increased to 62.5 mg/day of sunitinib in patients with good tolerance but in need for an increased anti-tumor activity, the plasma levels of sunitinib are remarkably similar in all patients irrespective of dose adaptation [Citation5]. These data suggest that individual patient characteristics that influence TKI absorption, excretion and metabolism may indeed influence TKI plasma levels and as a consequence determine the time and frequency of a dose reduction.
The efflux transporter ABCB1 (ATP binding cassette member B1, formerly known as P-glycoprotein or MDR1) is expressed in the intestine and liver and involved in the oral absorption and biliary secretion of several anticancer drugs [Citation15]. This transporter may contribute to multidrug resistance in tumors by actively extruding drugs from cancer cells, particularly in RCC [Citation16,Citation17]. As a consequence, expression levels and functionality of these drug transporters, i.e. due to polymorphisms, may have important consequences for the efficacy of sunitinib. The most common functional SNPs in ABCB1 are the synonymous 3435C > T (rs1045642) and 1236C > T (rs1128503) changes and the non-synonymous 2677G >T change (missense A893S/T rs2032582). Functional studies have shown that the haplotype of these three SNPs (rs1046542 – rs1128503 – rs2032582) alters the function of the efflux transporter including its substrate specificity. There are four publications showing an association between rs1128503 in ABCB1 and treatment outcome on anti-VEGFR-TKIs in mRCC () favoring patients with CT- and CC-variants [Citation7–9,Citation12]. As a consequence, the TT-genotype could lead to a more active efflux pump or more affinity of the pump for sunitinib, leading to lower sunitinib plasma levels. Our data suggesting an association between the TT-genotype and a delay in dose reductions supports this hypothesis.
Although Garcia-Donas, on a series of 89 mRCC patients treated with sunitinib, did not observe a higher risk for dose reductions in patients with the ABCB1 rs1128503 TT-variant or the rs2032582 TT/TA-variant, he observed less hypertension in patients with these variants: HR for the development hypertension was 0.41 (95% CI 0.20–0.81; p = 0.011) for the rs1128503 TT-variant and 0.42 (95% CI 0.21–0.84; p = 0.014) for the rs2032582 TT/TA-variant [Citation8]. Moreover, on a series of 115 mRCC patients treated with sunitinib, the TT-variant in rs2032582 in ABCB1 and the TT-variant of rs1128503 in ABCB1 were associated with a higher plasmatic sunitinib clearance (p = 0.02 and 0.05, respectively) [Citation18].
After absorption, sunitinib is converted to an equipotent metabolite, SU12662 [Citation19]. Both sunitinib and SU12662 are metabolized predominately by cytochrome (CYP) 3A4, and elimination is primarily via the feces. The expression of cytochrome CYP3A4, thought to be the key enzyme for the hepatic biotransformation of sunitinib, is regulated by the ligand-activated nuclear receptors NR1I2 (pregnane X receptor) and NR1I3 (constitutive androstane receptor) [Citation20,Citation21]. There is evidence that polymorphisms in NR1/2 and NR1/3 might be associated with outcome in mRCC treated with anti-VEGFR-TKIs (). Patients with the TT-genotype in rs2276707 in NR1/2, leading to a higher expression of CYP3A4, seem to have a shorter PFS and OS. Our findings of an association between the TT-genotype and a decreased delay in dose reductions support this hypothesis.
We could not find any association between the TT-variant in rs4073054 in NR1/3 and TTDR despite the fact that patients with this variant tend to have a worse outcome. Neither could we find any association between SNP rs776746 in CYP3A5 and TTDR, although it was shown that the AA- and AG-genotypes were link to improved treatment outcome [Citation7] and to increased need for dose reductions [Citation8].
These findings, when validated, could have interesting clinical applications. In fact, a patient whose disease is primarily or secondarily resistant to sunitinib 50 mg/day, who has few side effects and who has the ABCB1 rs1128503 TT-variant, the rs2032582 TT-variant or the NR1/2 rs2776707 TT-variant, could be a good candidate for a trial with sunitinib dose escalation to 62.5 mg/day or even 75 mg/day. There is evidence for the positive impact of dose escalation of some anti-VEGFR-TKIs on treatment outcome in mRCC. In a randomized phase II trial with sorafenib, dose escalation of sorafenib from 2 × 400 mg/d to 2 × 600 mg/d was foreseen. Forty-three (66%) of 65 patients who progressed on sorafenib 2 × 400 mg/d had their dose escalated and 42% of these patients achieved a reduction in tumor size and disease stabilization. The median PFS was 3.6 months for patients escalated to sorafenib 2 × 600 mg/d. The PFS of escalation of sorafenib was more effective than placebo in this setting [Citation22].
Our study has several potential limitations: 1) it was a retrospective analysis of patients treated in several centers without a central protocol dictating schedule and dose modifications or timing and method of radiological assessments; 2) the clinical sites did not report precise data on different side effects with National Cancer Institute Common Toxicity Criteria scoring, only the date of dose reduction and the reason for it were reported. Nevertheless, we assume that in most cases, the dose was reduced for grade 3 toxicity; 3) sunitinib plasma level were not available; 4) correction for multiple testing by Bonferroni, taking into account that the correlation with 11 SNPs was analyzed, indicated a p-value of < 0.005 as the threshold for significance. Our results did not reach this level of significance, probably due to the small number of patients in our series; 5) finally, there was better treatment outcome in our series (PFS 15.0 and OS 29.0 months) compared to the outcome on sunitinib in the pivotal trial (PFS 11.0 and OS 26.0 months [Citation1]). This difference is likely due to patient selection: all the patients had to complete at least one cycle of sunitinib and had to reach at least the first evaluation by CT scan.
Conclusion
Polymorphisms in the ABCB1 efflux pump are associated with the incidence of dose reductions in mRCC patients treated with sunitinib. Prospective validation of these findings including the association with sunitinib plasma levels is warranted and ongoing (EudraCT: 2011-006085-40/MetaSun).
Acknowledgments
This project is a common project of two kidney tumor banks: the CIT-rein tumor bank (Paris, France) and the University Hospitals Leuven kidney tumor bank (Leuven, Belgium). The CIT-rein project is headed by Professor Stéphane Oudard and Professor Jean-Jacques Patard. We want to thank sincerely for their collaboration the urologists, medical oncologists and pathologists of the following centers, whose biological material was used in the analysis: Angers: Centre Oncologique Paul Papin: Abdel Azzouzi, Rémy Delva, Stéphane Triau, Pierre Bigot; Créteil: Hôpital Henri Mondor: Alexandre de la Taille, Bernard Paule, Yves Allory; Suresnes: Hôpital Foch: Thierry Lebret, Christine Théodore, Yves Denoux; Leuven: University Hospitals Leuven: Hendrik Van Poppel, Evelyne Lerut, Joost Berkers, Pascal Wolter, Patrick Schöffski, Robert Paridaens; Limoges: Hôpital Dupuytren: Aurélien Descazeaud, Julien Berger; Lyon: Centre Léon Bérard: Marc Colombel, Sylvie Négrier, Florence Mege-Lechevallier; Marseille: Institut Paoli-Calmettes: Franck Bladou, Gwénaelle Gravis, Myriam Marcy; Nantes: ICO Gauducheau: Olivier Bouchot, Frédéric Rolland, Karine Reanudin; Paris: Hôpital Necker: Arnaud Méjean, Virginie Verkarre, Vincent Molinié; Poitiers: Jacques Irani, Jean Marc Tourani, Pierre Marie Le Villain; Rennes: Brigitte Laguerre, Jean-Jacques Patard, Nathalie Rioux-Leclercq; Strasbourg: CHRU Strasbourg: Didier Jacqmin, Brigitte Duclos, Véronique Lindler. The tissue collection was coordinated by the Plateforme de Ressources Biologiques de l’Hôpital Européen Georges Pompidou in Paris. We are grateful to Corine Takouchop Teghom, Claudia De Toma and Reza Elaidi for the coordination of the tissue and clinical data collection. Benoit Beuselinck received a grant from the Fondation Martine Midy (Paris, France) (2009–2010). His work is also funded by the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (Belgium) (2011–2013). Alexandra Karadimou received a grant from the Hellenic Society of Medical Oncology (Athens, Greece) (2010–2011). Diether Lambrechts is supported by the Stichting Tegen Kanker. Evelyne Lerut received fundings from Fonds voor Wetenschappelijk Onderzoek Vlaanderen (Belgium) and Stichting tegen Kanker. Stéphane Oudard received honorarium from Novartis, Pfizer, Roche and Bayer. Patrick Schöffski received research funding and honoriaria for consultant, advisory and educational functions from Pfizer and GSK. Benoit Beuselinck and Pascal Wolter are investigators of the EudraCT: 2011-006085-40/MetaSun trial financed by Pfizer. Benoit Beuselinck received honorarium from Bayer for educational activities. Pierre Bigot received honorarium from Novartis. Jean Jacques Patard is a consultant and principal investigator in Pfizer trials. The other authors have no conflicts of interest to declare.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med 2007;356: 115–24.
- Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Oudard S, et al. Overall survival and updated results for sunitinib compared with interferon alfa in patients with metastatic renal cell carcinoma. J Clin Oncol 2009;27:3584–90.
- Rini BI, Atkins MB. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol 2009;10:992–1000.
- Houk BE, Bello CL, Kang D, Amantea M. A population pharmacokinetic meta-analysis of sunitinib malate (SU11248) and its primary metabolite (SU12662) in healthy volunteers and oncology patients. Clin Cancer Res 2009;15:2497–506.
- Houk BE, Bello CL, Poland B, Rosen LS, Demetri GD, Motzer RJ. Relationship between exposure to sunitinib and efficacy and tolerability endpoints in patients with cancer: Results of a pharmacokinetic/pharmacodynamic meta-analysis. Cancer Chemother Pharmacol 2010;66:357–71.
- Rini BI, Cohen DP, Lu DR, Chen I, Hariharan S, Gore ME, et al. Hypertension as a biomarker of efficacy in patients with metastatic renal cell carcinoma treated with sunitinib. J Natl Cancer Inst 2011;103:763–73.
- van der Veldt AA, Eechoute K, Gelderblom H, Gietema J, Guchelaar HJ, van Erp NP, et al. Genetic polymorphisms associated with a prolonged progression-free survival in patients with metastatic renal cell cancer treated with sunitinib. Clin Cancer Res 2010;17:620–9.
- Garcia-Donas J, Esteban E, Leandro-García LJ, Castellano DE, Del Alba AG, Climent MA, et al. Single nucleotide polymorphism associations with response and toxic effects in patients with advanced renal-cell carcinoma treated with first-line sunitinib: A multicentre, observational, prospective study. Lancet Oncol 2011;12:1143–50.
- Beuselinck B, Karadimou A, Lambrechts D, Claes B, Wolter P, Couchy G, et al. Single nucleotide polymorphisms associated with outcome in metastatic renal cell carcinoma treated with sunitinib. Br J Cancer 2013;108:887–900.
- Beuselinck B, Karadimou A, Lambrechts D, Claes B, Wolter P, Couchy G, et al. VEGFR1 single nucleotide polymorphisms associated with outcome in patients with metastatic renal cell carcinoma treated with sunitinib – a multicentric retrospective analysis. Acta Oncol 2014;53: 103–12.
- Xu CF, Bing N, Ball H, Rajagopalan D, Sternberg C, Hutson T, et al. Pazopanib efficacy in renal cell carcinoma: Evidence for predictive genetic markers in angiogenesis- related and exposure-related genes. J Clin Oncol 2011; 29:2557–64.
- Xu CF, Ball HA, Bing N, Sternberg C, Xue Z, McCann L, et al. Association of genetic markers in angiogenesis- or exposure-related genes with overall survival in pazopanib-treated patients with advanced renal cell carcinoma. J Clin Oncol 2011;29(Suppl 7):abstr 303.
- Reumers J, De Rijk P, Zhao H, Liekens A, Smeets D, Cleary J, et al. Optimized filtering reduces the error rate in detecting genomic variants by short-read sequencing. J Nat Biotechnol 2011;30:61–8.
- Heng DY, Xie W, Regan MM et al. Prognostic factors for overall survival in patients with metastatic renal cell carcinoma treated with vascular endothelial growth factor-targeted agents: Results from a large, multicenter study. J Clin Oncol 2009;27:5794–9.
- Dietrich CG, Geier A, Oude Elferink RPJ. ABC of oral bioavailability: Transporters as gatekeepers in the gut. Gut 2003;52:1788–95.
- Walsh N, Larkin A, Kennedy S, Connolly L, Ballot J, Ooi W, et al. Expression of multidrug resistance markers ABCB1 (MDR-1/P-gp) and ABCC1 (MRP-1) in renal cell carcinoma. BMC Urol 2009;9:6.
- Soto-Vega E, Arroyo C, Richaud-Patin Y, Garcia-Carrasco M, Vazquez-Lavista LG, Llorente L. P-glycoprotein activity in renal clear cell carcinoma. Urol Oncol 2009;27: 363–6.
- Diekstra MH, Klümpen HJ, Lolkema MP, Yu H, Kloth JS, Gelderblom H, et al. Association Analysis of Genetic Polymorphisms in Genes Related to Sunitinib Pharmacokinetics, Specifically Clearance of Sunitinib and SU12662. Clin Pharmacol Ther 2014 Feb 24; doi: 10.1038/clpt.2014.47. [Epub ahead of print].
- Sakamoto KM. Su-11248 Sugen. Curr Opin Invest Drugs 2004;5:1329–39.
- Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, Lamba V, et al. The orphan nuclear receptor HNF4[alpha] determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat Med 2003;9:220–4.
- Kliewer SA, Goodwin B, Willson TM. The nuclear pregnane X receptor: A key regulator of xenobiotic metabolism. Endocr Rev 2002;23:687–702.
- Escudier B, Szczylik C, Hutson TE, Demkow T, Staehler M, Rolland F, et al. Randomized phase II trial of first-line treatment with sorafenib versus interferon alfa-2a in patients with metastatic renal cell carcinoma. J Clin Oncol 2009;27: 1280–9.