Abstract
Background. Optimal treatment decisions for cancer patients require reliable prognostic and predictive information. However, this information is inadequate in many cases. Several recent studies suggest that the leucine-rich repeats and immunoglobulin-like domains (LRIG) genes, transcripts, and proteins have prognostic implications in various cancer types.
Material and methods. Relevant literature was identified on PubMed using the key words lrig1, lrig2, and lrig3. LRIG mRNA expression in cancer versus normal tissues was investigated using the Oncomine database.
Results. The three human LRIG genes, LRIG1, LRIG2, and LRIG3, encode single-pass transmembrane proteins. LRIG1 is a negative regulator of growth factor signaling that has been shown to function as a tumor suppressor in vitro and in vivo in mice. The functions of LRIG2 and LRIG3 are less well defined. LRIG gene and protein expression are commonly dysregulated in human cancer. In early stage breast cancer, LRIG1 copy number was recently shown to predict early and late relapse in addition to overall survival; in nasopharyngeal carcinoma, loss of LRIG1 is also associated with poor survival. LRIG gene and protein expression have prognostic value in breast cancer, uterine cervical cancer, head-and-neck cancer, glioma, non-small cell lung cancer, prostate cancer, and cutaneous squamous cell carcinoma. In general, expression of LRIG1 and LRIG3 is associated with good survival, whereas expression of LRIG2 is associated with poor survival. Additionally, LRIG1 regulates cellular sensitivity to anti-cancer drugs, which indicates a possible role as a predictive marker.
Conclusions. LRIG gene statuses and mRNA and protein expression are clinically relevant prognostic indicators in several types of human cancer. We propose that LRIG analyses could become important when making informed and individualized clinical decisions regarding the management of cancer patients.
The need for new and better prognostic and predictive markers in clinical oncology is urgent because of the obvious over- and under-treatment of patients occurring today. One major explanation for suboptimal treatment decisions is the lack of prognostic and predictive markers that can be used to accurately predict disease recurrence and therapy response. The emerging tumor suppressor leucine-rich repeats and immunoglobulin-like domains- (LRIG-) 1 and its paralogs LRIG2 and LRIG3, have prognostic value in diverse types of cancer, including breast cancer [Citation1,Citation2], uterine cervical cancer [Citation3–5], head-and-neck cancer [Citation6–8], glioma [Citation9,Citation10], prostate cancer [Citation11], and cutaneous squamous cell carcinoma [Citation12]. A brief mini-review on LRIG in cancer was recently published [Citation13]; however, no thorough overview of the clinical implications of the many new and important findings in the LRIG field have been presented since our previous LRIG review in 2007 [Citation14]. Here, we summarize the current knowledge on LRIG and cancer prognosis and discuss the potential future role of LRIG genes and proteins as clinically useful molecular markers in human cancer.
Material and methods
Literature survey
PubMed (http://www.ncbi.nlm.nih.gov/pubmed) was searched using the search terms lrig1, lrig2, and lrig3, which yielded 109, 25, and 29 entries, respectively, of which 107 were non-redundant, peer-reviewed original articles written in the English language. Of these articles, 17 contained both LRIG expression data and patient survival data, and 11 presented significant up- or down-regulation of LRIG1 mRNA or protein in cancer tissue compared with normal tissue; all of these articles were included in the present review. Additionally, key papers describing the identification and molecular, developmental, and physiological functions of the LRIG proteins were included.
Analysis of LRIG mRNA expression in different cancer types
The possible differences in LRIG mRNA expression in cancer versus normal tissues was investigated using the Oncomine database (Compendia Bioscience, Ann Arbor, MI, USA) with Oncomine 4.4.3 Research Edition with the following settings: threshold (p-value), 10−4; threshold (fold change), all; threshold (gene rank), all. For each cancer type, changes in expression were reported as over- or under-expression if they were found in two or more data sets.
Results
The LRIG family
The human LRIG gene family comprises three genes, LRIG1, LRIG2, and LRIG3, which are located at chromosomes 3p14, 1p13.1, and 12q14.1, respectively [Citation15–17]. The encoded trans-membrane proteins share a similar structure with an extra-cellular or luminal leucine-rich-repeat domain and three immunoglobulin-like domains, a transmembrane domain, and a cytosolic domain. All three LRIG genes and proteins are widely expressed in human and mouse tissues [Citation15–20]. The subcellular localization of LRIG proteins varies and includes the plasma membrane, cytoplasm, perinuclear region, and nucleus [Citation14]. However, in certain pathological tissues, such as psoriatic skin, the subcellular distributions of the LRIG proteins are altered [Citation21].
LRIG1 is an emerging tumor suppressor
More than 10 years ago, LRIG1 was hypothesized to be a tumor suppressor gene [Citation15,Citation22]. Experimental and clinical data have subsequently supported this original hypothesis. Accumulating evidence shows that LRIG1 suppresses the proliferation of normal and transformed cells. Cells whose proliferation is inhibited by LRIG1 include human embryonic kidney cells [Citation23], prostate cancer cells [Citation11,Citation24], mouse fibroblasts [Citation24], keratinocytes [Citation25], bladder cancer cells [Citation26], breast cancer cells [Citation1,Citation27,Citation28], astrocytoma and glioblastoma cells [Citation29,Citation30], African green monkey kidney cells [Citation30], nasopharyngeal carcinoma cells [Citation7], and lung cancer cells [Citation31]. At the molecular level, LRIG1 negatively regulates growth factor signaling mediated by several oncogenic receptor tyrosine kinases. This appears to be achieved through different mechanisms: LRIG1 induces the ubiquitylation and degradation of the epidermal growth factor receptor (EGFR) family members EGFR, ERBB2 (also called HER2), ERBB3, and ERBB4 [Citation23,Citation24], destabilizes the MET receptor [Citation27,Citation32], and physically interacts with the RET receptor to inhibit RET phosphorylation and downstream signaling [Citation33]. The platelet-derived growth factor (PDGF) receptor PDGFRα is down-regulated by LRIG1 through an as-yet undefined mechanism [Citation34]. Furthermore, LRIG1 also appears to be able to regulate cytokine and bone morphogenetic protein (BMP) signaling; Lrig1-deficient cells show an increased responsiveness to interleukin-6 [Citation35], and SMA-10/LRIG mutant nematodes show impaired BMP signaling [Citation36]. In addition to the cell-autonomous functions, LRIG1 ectodomains can be shed to negatively regulate EGFR signaling in a paracrine manner [Citation30,Citation37]. Thus, several mechanisms for the LRIG1 protein that negatively regulates receptor signaling have been described thus far, and the non-cell- autonomous inhibition of growth factor receptor signaling via soluble LRIG1 ectodomains could be a mechanism that could be exploited for anti-cancer treatment [Citation30,Citation37,Citation38]. Recently, Lrig1 was proven to function as a tumor suppressor in mice. Lrig1- deficient mice show hyperproliferation of epidermal [Citation25,Citation39,Citation40] and intestinal [Citation41,Citation42] stem cells, airway hyperplasia [Citation31], and development of spontaneous intestinal adenomas [Citation42].
The role of LRIG2 and LRIG3 in cancer
LRIG2 and LRIG3 are less studied than LRIG1. Recently, it was found that a subgroup of individuals with urofacial syndrome carries mutations in LRIG2 [Citation43]. Lrig2-deficient mice display transiently reduced growth and increased spontaneous mortality [Citation34]. Intriguingly and in contrast with LRIG1, LRIG2 may promote gliomagenesis, as Lrig2-deficient mice are protected against PDGF-induced glioma [Citation34]. This finding is consistent with results showing that the down-regulation of LRIG2 in a human glioma cell line results in decreased cell proliferation and increased apoptosis [Citation44]. Lrig3-deficient mice show disturbed development of the inner ear and present with craniofacial defects [Citation45,Citation46]. In the amphibian Xenopus laevis, LRIG3 inhibits fibroblast growth factor (FGF) signaling [Citation47]. Recently, mammalian LRIG3 was proposed to oppose the function of LRIG1, thus stabilizing the EGFR family proteins [Citation48]. Despite these intriguing observations, the roles of LRIG2 and LRIG3 in human cancer remain poorly understood.
LRIG expression is dysregulated in human cancer
All of the LRIG genes and proteins are expressed in most, if not all, normal human tissues [Citation15–18]. However, relatively little is known about the regulation of LRIG expression. The transcription of LRIG1 can be induced using growth factors [Citation23] or steroid hormones [Citation1,Citation11]. In ERBB2-positive cancer cell lines, over-expression of ERBB2 down-regulates the estrogen receptor, thereby suppressing the estrogen-induced transcription of LRIG1 [Citation1,Citation28]. The natural compound gambogic acid up-regulates LRIG1 in glioma cells through the activation of AMP-regulated kinase, thereby inhibiting the proliferation of these cells [Citation49]. In the lung, LRIG1 appears to be down-regulated by smoking [Citation50], which is consistent with the observation that in early stage uterine cervical tumors, LRIG1 levels are lower in smoking women than in non-smoking women [Citation3]. The transcription of LRIG2 has been demonstrated to be regulated by the concomitant expression of the transcription factors FOS, TAF1, and YY1 [Citation51]. This could potentially be relevant in cancer biology because FOS is a well-established proto-oncogene. Expression of Lrig3 is regulated by Netrin during mouse embryonic inner ear lateral canal development [Citation45]; however, the significance of the regulation of LRIG3 in other organs and in neoplasia is unknown.
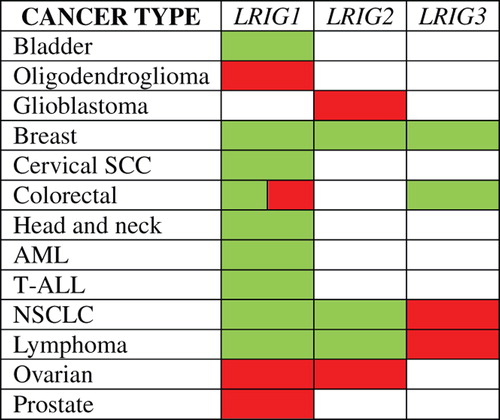
Given that the LRIG proteins are important regulators of growth factor signaling and cell proliferation, it is not surprising that their expression is often dysregulated in human cancer. For example, it was recently demonstrated that down-regulation of LRIG1 is associated with the benign-to-malignant transformation of ocular-surface squamous tumors [Citation52]. Additionally, LRIG1 mRNA and protein have been reported to be significantly down-regulated in neoplastic tissue compared with the corresponding normal tissue in breast cancer and ERBB2-positive breast cancer [Citation28], clear cell renal cell carcinoma [Citation53], chronic lymphocytic leukemia [Citation54], oral squamous cell carcinoma [Citation55], astrocytoma [Citation56], lung squamous cell carcinoma and carcinoma in situ [Citation31,Citation50], nasopharyngeal squamous cell carcinoma [Citation7], and colorectal cancer [Citation13]. However, LRIG1 mRNA and protein are significantly over-expressed in prostate cancer [Citation11] and breast cancer of the luminal A subtype [Citation13], which are two cancer types that are largely driven by steroid hormones. An analysis of the gene expression data sets at Oncomine confirmed that LRIG genes are aberrantly expressed in many neoplastic tissues compared with their normal tissue counterparts. In particular, dysregulated LRIG expression was found in cancers of the bladder, breast, uterine cervix, colorectum, head-and-neck, hematopoietic system, ovary, and prostate and also in glioma (). Notably, this list derived from Oncomine likely displays only a fraction of all of the cancer types with dysregulated LRIG expression because stringent criteria for over- and under-expression were used in the analysis (see Material and methods). The biological interpretation of dysregulated LRIG expression in cancer tissues is, however, complicated by the fact that expression is physiologically regulated and may be selected for (or against) during tumor evolution [Citation11]. That is, it cannot be a priori deduced whether a given LRIG gene or protein will be over- or under-expressed in a certain cancer type at a specific time point. Additionally, genomic aberrations of LRIG genes are frequently found. In nasopharyngeal carcinoma, LRIG1 is commonly deleted [Citation6], and in breast cancer, LRIG1 shows both increased and decreased gene copy numbers, and gene recombination events have been suggested [Citation2,Citation57,Citation58].
Figure 1. Over- and under-expression of LRIG genes in human cancers compared with their normal tissue counterparts. Altered expression is indicated only when two or more studies in the Oncomine database showed over- or under-expression for their respective cancer type. Red indicates over-expression and green indicates under-expression in tumors versus normal tissue. Colorectal cancer showed both over- and under-expression of LRIG1. AML, acute myeloid leukemia; NSCLC, non-small cell lung cancer; SCC, squamous cell carcinoma; T-ALL, T-cell acute lymphoblastic leukemia.
LRIG as prognostic indicators in human cancer
Loss of LRIG1 predicts early and late relapse in breast cancer
Late relapse in breast cancer remains an important clinical problem. Currently, there are very few, if any, molecular markers that robustly predict the risk of late relapse in early stage breast cancer. A recent study of gene copy number variation in 971 stage I/II breast tumors revealed that loss of the LRIG1 locus is an independent risk factor for early and late relapse and for death [Citation2]. In another recent study, LRIG1 expression was higher in patients with tumors expressing the estrogen receptor, and data collected from Oncomine showed that high expression of LRIG1 in estrogen receptor-positive and lymph node-negative breast cancer was correlated with prolonged relapse-free survival [Citation1]. Given that LRIG1 negatively regulates the ERBB2 receptor and the proliferation of ERBB2-positive breast cancer cells, it will be interesting to investigate whether LRIG1 also has prognostic value in ERBB2-positive breast cancer. In any case, if the LRIG1 copy number or expression level is confirmed to predict late relapse in early stage breast cancer, LRIG1 could become a clinically relevant molecular classifier in breast cancer.
In uterine cervical cancer, expression of LRIG1 and LRIG2 is associated with good and poor prognosis, respectively
LRIG expression was studied via immunohistochemistry in 128 women with uterine cervical cancer [Citation3,Citation4] and 86 women with uterine adenocarcinoma [Citation5]. In cervical squamous cell carcinoma, both LRIG1 and LRIG2 were independent prognostic factors. Intriguingly, in early stage squamous cell carcinoma, high expression of LRIG1 correlated with good survival [Citation3], whereas high expression of LRIG2 correlated with poor survival [Citation4]. When low expression of LRIG1 and high expression of LRIG2 were combined, a subgroup of patients with a very poor prognosis was identified [Citation4], which suggests that LRIG1 and LRIG2 could be useful prognostic markers in early stage uterine squamous cell carcinoma; it also suggests that LRIG1 and LRIG2 could have different and opposing functions in which LRIG1 suppresses cancer and LRIG2 promotes cancer. No correlation between LRIG expression and survival was found in cervical squamous cell carcinoma at later stages [Citation3]. In uterine cervical adenocarcinoma, expression of both LRIG1 and LRIG3 correlated with good survival [Citation5]. When all the significant parameters, LRIG1, LRIG3, HPV status, age, histology, and FIGO stage were included in a multivariate analysis, only LRIG3 and age showed independent statistical significance [Citation5]. In summary, these studies indicate that LRIG proteins could be relevant prognostic markers in cervical cancers of different histologies and for patients of different stages.
Head-and-neck cancer
In nasopharyngeal carcinoma, the LRIG1 locus is frequently deleted and the mRNA levels decreased [Citation6]. In a panel of 48 clinical samples, 30% showed a loss of LRIG1, and concomitant loss of LRIG1 and ADAMTS9 was associated with a poor clinical outcome. More recently, an immunohistochemical analysis of 44 metastatic nasopharyngeal carcinomas revealed a significant correlation between low LRIG1 expression and poor outcome [Citation7]. Similarly, in oropharyngeal carcinoma, an immunohistochemical study of 278 cases showed that LRIG1 was an independent positive prognostic factor [Citation8]. Of note, the subgroup of patients with oropharyngeal tumors that were human papilloma virus-positive and expressed high LRIG1 levels showed a very favorable prognosis. These criteria may possibly define a subgroup of patients where adjuvant treatment is not necessary, whereas patients with LRIG1-negative tumors could comprise a subgroup that could benefit from intensified treatment. Together, these studies indicate that LRIG1 could be a clinically useful prognostic marker in head-and-neck cancer.
Perinuclear expression of LRIG proteins is associated with prolonged survival in astrocytic tumors
The prognostic impact of the expression of LRIG proteins was investigated by immunohistochemistry in 404 patients with astrocytic brain tumors [Citation9]. Perinuclear immunoreactivity of all LRIG proteins was inversely correlated with World Health Organization (WHO) grade, which is a known prognostic factor in astrocytic tumors. Additionally, perinuclear and cytoplasmic staining of LRIG3 was correlated with a lower proliferation index, and perinuclear staining and WHO tumor grade were independent prognostic factors in the analyzed cohort. Regrettably, the currently established prognostic and predictive molecular markers in glioma, MGMT promoter methylation, chromosome 1p and 19q co-deletion, and IDH1 mutations were not analyzed in this patient series, thereby precluding a comparison between LRIG3 and these markers. Nevertheless, the data suggest that the expression of the LRIG proteins and in particular perinuclear expression of LRIG3 could be important when evaluating the prognosis of astrocytic tumors.
Expression of LRIG2 predicts poor survival in oligodendroglial tumors
In an immunohistochemical study including 63 patients with oligodendroglial brain tumors, LRIG expression was assessed in relation to various clinical parameters and patient survival [Citation10]. Similar to the situation in early stage cervical squamous cell carcinoma, LRIG2 was an independent prognostic factor in oligodendroglial tumors. Unfortunately, neither in this glioma series were MGMT promoter methylation status, chromosome 1p and 19q co-deletion, nor IDH1 mutations analyzed. However, a subgroup analysis suggested that there was no association between LRIG2 expression and the numerical status of chromosome 1p. The finding that LRIG2 expression was associated with poor oligodendroglioma patient survival was further corroborated in an experimental study showing a correlation between Lrig2 gene dosage and the incidence and malignancy of PDGFB-induced glioma [Citation34]. Thus, in both cervical and oligodendroglial cancer, expression of LRIG2 predicts poor survival, emphasizing that the function of LRIG2 may be different from that of LRIG1.
Expression of LRIG1 is associated with good survival in non-small cell lung cancer
LRIG1 is down-regulated in non-small cell lung cancer and lung carcinoma in situ [Citation14,Citation31,Citation50] (Kvarnbrink et al., unpublished observation). Both the mRNA and protein levels of LRIG1 are associated with the survival of non-small cell lung cancer patients. In an immunohistochemical analysis of 347 non-small cell lung cancer cases, Kvarnbrink and co-workers found that high expression of LRIG1 was an independent prognostic factor that was correlated with good survival, particularly among patients with lung adenocarcinoma (Kvarnbrink et al., unpublished observation). Intriguingly, in lung squamous cell carcinoma, expression of LRIG1 was not associated with survival if only the neoplastic cells were analyzed, whereas if the stromal and immune cells were also included in the analysis, high LRIG1 expression was correlated with a better survival. In the Oncomine database, which includes composite gene expression profiles of all cell types included in the respective tumor specimens, the expression of LRIG1 mRNA was associated with good survival in both lung adenocarcinoma and lung squamous cell carcinoma. Thus, LRIG1 expression in cancer epithelial cells and in stromal and immune cells could provide prognostic information in non-small cell lung cancer. Taken together, these findings indicate that LRIG1 could be an important prognostic factor in the two major types of non-small cell lung cancer, i.e. lung adenocarcinoma and lung squamous cell carcinoma. Furthermore, LRIG1 immunoreactivity predicted survival in a cohort that was primarily composed of early stage cases, a group of patients where new prognostic markers are especially needed.
Expression of LRIG1 shows different prognostic value depending on the clinical context in prostate cancer
Expression of LRIG1 was evaluated in a Swedish cohort of 355 men and in an American cohort of 293 men via the immunohistochemical staining of tissue microarrays [Citation11]. The two cohorts had different clinical features and showed divergent results. In the Swedish cohort, the patients were followed via watchful waiting, and high expression of LRIG1 was an independent prognostic factor that was associated with worse overall and disease-specific survival. The American cohort included patients who had undergone prostatectomy; in this group, the expression of LRIG1 was a positive prognostic factor for overall survival. The clinical and biological circumstances are therefore important determinants for the prognostic value of LRIG1 expression in prostate cancer.
LRIG1 expression is associated with good survival in cutaneous squamous cell carcinoma
In cutaneous squamous cell carcinoma, LRIG1 expression was investigated via immunohistochemistry in a series of 38 patients [Citation12]. High expression of LRIG1 correlated with a higher grade of differentiation, the absence of regional or distant metastasis, and better overall survival. However, it was not investigated whether LRIG1 was an independent prognostic marker in this cohort or whether its association with survival was dependent on its correlation with histological grade or the absence of metastases.
Prognostic impact of LRIG protein and mRNA expression in other cancer types
In one study, expression of LRIG1 was evaluated in colorectal cancer via immunoblotting of the tumor and non-neoplastic tissue lysates from 30 patients [Citation59]. LRIG1 was both up- and down-regulated; however, the available data were too limited to perform a meaningful survival analysis. As LRIG1 is a tumor suppressor in the mouse intestine, further studies to evaluate the prognostic implications of LRIG expression in human colorectal cancer may be warranted.
In a series of 31 renal cell carcinomas, LRIG1 and EGFR expression was analyzed via quantitative real-time RT-PCR [Citation60]. The LRIG1/EGFR ratio was reduced in renal cell carcinoma tissue compared with normal kidney cortex. In a follow-up study including 104 renal cell carcinoma cases [Citation53], LRIG1 expression was down-regulated in renal cell carcinoma; however, this was observed only in the clear cell subtype. There was no correlation between LRIG1 expression and patient survival in this cohort.
In esophageal cancer, the prognostic value of the immunohistochemical expression of different LRIG proteins in tumors from 80 patients was evaluated; no significant correlations between LRIG expression and patient survival were found [Citation61].
Ghasimi and co-workers analyzed LRIG expression in 409 meningioma cases via immunohistochemistry [Citation62]. Expression of both LRIG1 and LRIG2 correlated with estrogen receptor status, which is consistent with the finding that LRIG1 is regulated by estrogen signaling in breast cancer [Citation1]. Because meningioma is usually benign, the vast majority of patients in Ghasimi's study did not succumb to their disease. Therefore, it was not surprising that no association between LRIG protein expression and meningioma patient survival was observed in this study.
A gene expression meta-analysis supports a major role for LRIG1 as a prognostic indicator in additional cancer types
In a meta-analysis of gene expression and patient outcome in nine studies of breast, lung, and bladder cancer, glioma and melanoma, LRIG1 was identified as one of the four top genes that predicted survival across all of the data sets (the other genes were, KIF4A, HJURP, and ESPL1); the high expression of LRIG1 was associated with a better prognosis for these patients [Citation63]. This finding supports the results discussed above for breast and lung cancer, and glioma, and it also suggests that LRIG1 expression could have prognostic value in additional cancer types including bladder cancer and melanoma. A summary of all published articles showing significant associations between LRIG gene status, mRNA expression, or protein expression and cancer patient survival is presented in .
Table I. Correlation between LRIG1 gene copy number status, LRIG1 mRNA expression, or LRIG protein expression and patient survival in various cancer types.
LRIG1 as a predictive marker of therapy response
LRIG1 predicts and regulates chemotherapy responses in cancer cell lines
In a series of nine esophageal cancer cell lines, expression of LRIG1 was correlated with sensitivity to the chemotherapeutic agents cisplatin, docetaxel, and vinorelbine [Citation61]. Consistent with this finding, ectopic expression of LRIG1 increases the sensitivity of bladder cancer cells to cisplatin [Citation64] and of glioblastoma cells to cisplatin and temozolomide [Citation29,Citation65]. Additionally, LRIG1 confers sensitivity of breast cancer cells to an experimental Smac mimetic inducer of apoptosis [Citation66] and of gastric carcinoma cells to a MET-targeting antibody [Citation32]. Taken together, these and other results indicate that LRIG1 could have potential both as a therapeutic agent and as a predictive marker. In some of the studies referred to above, the prognostic value of LRIG1 expression may have reflected the effects of LRIG1 on the sensitivity of the tumors to the respective therapies used. Determination of whether the effects of LRIG expression on patient survival are truly prognostic or predictive will require patient materials from controlled clinical studies including different treatment arms. The finding that LRIG1 could serve as a predictive marker should be further investigated because there is a great need for predictive biomarkers because of the rapid increase of treatment alternatives in medical oncology.
Conclusions
The expression and gene status of LRIG1, LRIG2, and LRIG3 have prognostic and possibly predictive value in diverse types of cancer. These markers may assist in settings of early stage cancers where current treatment decisions are often made in the absence of adequate prognostic and predictive information. It will be important to establish methods to analyze LRIG expression and LRIG gene status in the clinical setting and to investigate the clinical value of LRIG analyses in retrospective clinical trials with the view that LRIG analyses could become important when making informed and individualized clinical decisions regarding the management of cancer patients.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Krig SR, Frietze S, Simion C, Miller JK, Fry WH, Rafidi H, et al. Lrig1 is an estrogen-regulated growth suppressor and correlates with longer relapse-free survival in ER alpha-positive breast cancer. Mol Cancer Res 2011;9: 1406–17.
- Thompson PA, Ljuslinder I, Tsavachidis S, Brewster A, Sahin A, Hedman H, et al. Loss of LRIG1 locus increases risk of early and late relapse of stage I/II breast cancer. Cancer Res 2014;74:2928–35.
- Lindstrom AK, Ekman K, Stendahl U, Tot T, Henriksson R, Hedman H, et al. LRIG1 and squamous epithelial uterine cervical cancer: Correlation to prognosis, other tumor markers, sex steroid hormones, and smoking. Int J Gynecol Cancer 2008;18:312–7.
- Hedman H, Lindstrom AK, Tot T, Stendahl U, Henriksson R, Hellberg D. LRIG2 in contrast to LRIG1 predicts poor survival in early-stage squamous cell carcinoma of the uterine cervix. Acta Oncol 2010;49:812–5.
- Muller S, Lindquist D, Kanter L, Flores-Staino C, Henriksson R, Hedman H, et al. Expression of LRIG1 and LRIG3 correlates with human papillomavirus status and patient survival in cervical adenocarcinoma. Int J Oncol 2013;42:247–52.
- Sheu JJ, Lee CH, Ko JY, Tsao GS, Wu CC, Fang CY, et al. Chromosome 3p12.3-p14.2 and 3q26.2-q26.32 are genomic markers for prognosis of advanced nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev 2009;18: 2709–16.
- Sheu JJ, Lee CC, Hua CH, Li CI, Lai MT, Lee SC, et al. LRIG1 modulates aggressiveness of head and neck cancers by regulating EGFR-MAPK-SPHK1 signaling and extracellular matrix remodeling. Oncogene 2014;33:1375–84.
- Lindquist D, Näsman A, Tarján M, Henriksson R, Tot T, Dalianis T, et al. Expression of LRIG1 is associated with good prognosis and human papillomavirus status in oropharyngeal cancer. Br J Cancer 2014;110:1793–800.
- Guo D, Nilsson J, Haapasalo H, Raheem O, Bergenheim T, Hedman H, et al. Perinuclear leucine-rich repeats and immunoglobulin-like domain proteins (LRIG1-3) as prognostic indicators in astrocytic tumors. Acta Neuropathol 2006; 111:238–46.
- Holmlund C, Haapasalo H, Yi W, Raheem O, Brannstrom T, Bragge H, et al. Cytoplasmic LRIG2 expression is associated with poor oligodendroglioma patient survival. Neuropathology 2009;29:242–7.
- Thomasson M, Wang B, Hammarsten P, Dahlman A, Persson JL, Josefsson A, et al. LRIG1 and the liar paradox in prostate cancer: A study of the expression and clinical significance of LRIG1 in prostate cancer. Int J Cancer 2011;128:2843–52.
- Tanemura A, Nagasawa T, Inui S, Itami S. LRIG-1 provides a novel prognostic predictor in squamous cell carcinoma of the skin: Immunohistochemical analysis for 38 cases. Dermatol Surg 2005;31:423–30.
- Wang Y, Poulin EJ, Coffey RJ. LRIG1 is a triple threat: ERBB negative regulator, intestinal stem cell marker and tumour suppressor. Br J Cancer 2013;108:1765–70.
- Hedman H, Henriksson R. LRIG inhibitors of growth factor signalling – double-edged swords in human cancer? Eur J Cancer 2007;43:676–82.
- Nilsson J, Vallbo C, Guo D, Golovleva I, Hallberg B, Henriksson R, et al. Cloning, characterization, and expression of human LIG1. Biochem Biophys Res Commun 2001;284:1155–61.
- Holmlund C, Nilsson J, Guo D, Starefeldt A, Golovleva I, Henriksson R, et al. Characterization and tissue-specific expression of human LRIG2. Gene 2004;332:35–43.
- Guo D, Holmlund C, Henriksson R, Hedman H. The LRIG gene family has three vertebrate paralogs widely expressed in human and mouse tissues and a homolog in Ascidiacea. Genomics 2004;84:157–65.
- Nilsson J, Starefeldt A, Henriksson R, Hedman H. LRIG1 protein in human cells and tissues. Cell Tissue Res 2003; 312:65–71.
- Suzuki Y, Sato N, Tohyama M, Wanaka A, Takagi T. cDNA cloning of a novel membrane glycoprotein that is expressed specifically in glial cells in the mouse brain. LIG-1, a protein with leucine-rich repeats and immunoglobulin-like domains. J Biol Chem 1996;271:22522–7.
- Homma S, Shimada T, Hikake T, Yaginuma H. Expression pattern of LRR and Ig domain-containing protein (LRRIG protein) in the early mouse embryo. Gene Expr Patterns 2009;9:1–26.
- Karlsson T, Mark EB, Henriksson R, Hedman H. Redistribution of LRIG proteins in psoriasis. J Invest Dermatol 2008; 128:1192–5.
- Hedman H, Nilsson J, Guo D, Henriksson R. Is LRIG1 a tumour suppressor gene at chromosome 3p14.3? Acta Oncol 2002;41:352–4.
- Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. EMBO J 2004;23: 3270–81.
- Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, 3rd, et al. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem 2004;279:47050–6.
- Jensen KB, Watt FM. Single-cell expression profiling of human epidermal stem and transit-amplifying cells: Lrig1 is a regulator of stem cell quiescence. Proc Natl Acad Sci U S A 2006;103:11958–63.
- Yang WM, Yan ZJ, Ye ZQ, Guo DS. LRIG1, a candidate tumour-suppressor gene in human bladder cancer cell line BIU87. Br J Urol Int 2006;98:898–902.
- Shattuck DL, Miller JK, Laederich M, Funes M, Petersen H, Carraway KL, 3rd, et al. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Mol Cell Biol 2007;27:1934–46.
- Miller JK, Shattuck DL, Ingalla EQ, Yen L, Borowsky AD, Young LJ, et al. Suppression of the negative regulator LRIG1 contributes to ErbB2 overexpression in breast cancer. Cancer Res 2008;68:8286–94.
- Stutz MA, Shattuck DL, Laederich MB, Carraway KL 3rd, Sweeney C. LRIG1 negatively regulates the oncogenic EGF receptor mutant EGFRvIII. Oncogene 2008;27:5741–52.
- Yi W, Holmlund C, Nilsson J, Inui S, Lei T, Itami S, et al. Paracrine regulation of growth factor signaling by shed leucine-rich repeats and immunoglobulin-like domains 1. Exp Cell Res 2011;317:504–12.
- Lu L, Teixeira VH, Yuan Z, Graham TA, Endesfelder D, Kolluri K, et al. LRIG1 regulates cadherin-dependent contact inhibition directing epithelial homeostasis and pre-invasive squamous cell carcinoma development. J Pathol 2013;229:608–20.
- Lee JM, Kim B, Lee SB, Jeong Y, Oh YM, Song YJ, et al. Cbl-independent degradation of Met: Ways to avoid agonism of bivalent Met-targeting antibody. Oncogene 2014;33:34–43.
- Ledda F, Bieraugel O, Fard SS, Vilar M, Paratcha G. Lrig1 is an endogenous inhibitor of Ret receptor tyrosine kinase activation, downstream signaling, and biological responses to GDNF. J Neurosci 2008;28:39–49.
- Rondahl V, Holmlund C, Karlsson T, Wang B, Faraz M, Henriksson R, et al. Lrig2-deficient mice are protected against PDGFB-induced glioma. PLoS One 2013;8:e73635.
- Nakamura T, Hamuro J, Takaishi M, Simmons S, Maruyama K, Zaffalon A, et al. LRIG1 inhibits STAT3-dependent inflammation to maintain corneal homeostasis. J Clin Invest 2014;124:385–97.
- Gumienny TL, Macneil L, Zimmerman CM, Wang H, Chin L, Wrana JL, et al. Caenorhabditis elegans SMA-10/LRIG is a conserved transmembrane protein that enhances bone morphogenetic protein signaling. PLoS Genet 2010; 6:e1000963.
- Goldoni S, Iozzo RA, Kay P, Campbell S, McQuillan A, Agnew C, et al. A soluble ectodomain of LRIG1 inhibits cancer cell growth by attenuating basal and ligand-dependent EGFR activity. Oncogene 2007;18;26:368–81.
- Johansson M, Oudin A, Tiemann K, Bernard A, Keunen O, Fack F, et al. The soluble form of the tumor suppressor Lrig1 potently inhibits in vivo glioma growth irrespective of EGF receptor status. Neuro Oncol 2013;15:1200–11.
- Suzuki Y, Miura H, Tanemura A, Kobayashi K, Kondoh G, Sano S, et al. Targeted disruption of LIG-1 gene results in psoriasiform epidermal hyperplasia. FEBS Lett 2002;521: 67–71.
- Jensen KB, Collins CA, Nascimento E, Tan DW, Frye M, Itami S, et al. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 2009;4:427–39.
- Wong VW, Stange DE, Page ME, Buczacki S, Wabik A, Itami S, et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol 2012;14:401–8.
- Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 2012;149:146–58.
- Stuart HM, Roberts NA, Burgu B, Daly SB, Urquhart JE, Bhaskar S, et al. LRIG2 mutations cause urofacial syndrome. Am J Hum Genet 2013;92:259–64.
- Wang B, Han L, Chen R, Cai M, Han F, Lei T, et al. Downregulation of LRIG2 expression by RNA interference inhibits glioblastoma cell (GL15) growth, causes cell cycle redistribution, increases cell apoptosis and enhances cell adhesion and invasion in vitro. Cancer Biol Ther 2009;8: 1018–23.
- Abraira VE, Del Rio T, Tucker AF, Slonimsky J, Keirnes HL, Goodrich LV. Cross-repressive interactions between Lrig3 and netrin 1 shape the architecture of the inner ear. Development 2008;135:4091–9.
- Abraira VE, Satoh T, Fekete DM, Goodrich LV. Vertebrate Lrig3-ErbB interactions occur in vitro but are unlikely to play a role in Lrig3-dependent inner ear morphogenesis. PLoS One 2010;5:e8981.
- Zhao H, Tanegashima K, Ro H, Dawid IB. Lrig3 regulates neural crest formation in Xenopus by modulating Fgf and Wnt signaling pathways. Development 2008;135: 1283–93.
- Rafidi H, Mercado F 3rd, Astudillo M, Fry WH, Saldana M, Carraway KL 3rd, et al. Leucine-rich repeat and immunoglobulin domain-containing protein-1 (Lrig1) negative regulatory action toward ErbB receptor tyrosine kinases is opposed by leucine-rich repeat and immunoglobulin domain-containing protein 3 (Lrig3). J Biol Chem 2013;288: 21593–605.
- He XY, Liu XJ, Chen X, Bian LG, Zhao WG, Shen JK, et al. Gambogic acid induces EGFR degradation and Akt/mTORC1 inhibition through AMPK dependent-LRIG1 upregulation in cultured U87 glioma cells. Biochem Biophys Res Commun 2013;435:397–402.
- Boelens MC, van den Berg A, Fehrmann RS, Geerlings M, de Jong WK, te Meerman GJ, et al. Current smoking-specific gene expression signature in normal bronchial epithelium is enhanced in squamous cell lung cancer. J Pathol 2009; 218:182–91.
- Wang D, Rendon A, Ouwehand W, Wernisch L. Transcription factor co-localization patterns affect human cell type-specific gene expression. BMC Genomics 2012;13:263.
- Nagata M, Nakamura T, Sotozono C, Inatomi T, Yokoi N, Kinoshita S. LRIG1 as a potential novel marker for neoplastic transformation in ocular surface squamous neoplasia. PLoS One 2014;9:e93164.
- Thomasson M, Hedman H, Ljungberg B, Henriksson R. Gene expression pattern of the epidermal growth factor receptor family and LRIG1 in renal cell carcinoma. BMC Res Notes 2012;5:216.
- Hanlon K, Rudin CE, Harries LW. Investigating the targets of MIR-15a and MIR-16-1 in patients with chronic lymphocytic leukemia (CLL). PLoS One 2009;4:e7169.
- Jensen KB, Jones J, Watt FM. A stem cell gene expression profile of human squamous cell carcinomas. Cancer Lett 2008;272:23–31.
- Ye F, Gao Q, Xu T, Zeng L, Ou Y, Mao F, et al. Upregulation of LRIG1 suppresses malignant glioma cell growth by attenuating EGFR activity. J Neurooncol 2009;94:183–94.
- Ljuslinder I, Malmer B, Golovleva I, Thomasson M, Grankvist K, Höckenström T, et al. Increased copy number at 3p14 in breast cancer. Breast Cancer Res 2005;7:R719–27.
- Ljuslinder I, Golovleva I, Henriksson R, Grankvist K, Malmer B, Hedman H. Co-incidental increase in gene copy number of ERBB2 and LRIG1 in breast cancer. Breast Cancer Res 2009;11:403.
- Ljuslinder I, Golovleva I, Palmqvist R, Oberg A, Stenling R, Jonsson Y, et al. LRIG1 expression in colorectal cancer. Acta Oncol 2007;46:1118–22.
- Thomasson M, Hedman H, Guo D, Ljungberg B, Henriksson R. LRIG1 and epidermal growth factor receptor in renal cell carcinoma: A quantitative RT-PCR and immunohistochemical analysis. Br J Cancer 2003;89:1285–9.
- Wu X, Hedman H, Bergqvist M, Bergstrom S, Henriksson R, Gullbo J, et al. Expression of EGFR and LRIG proteins in oesophageal carcinoma with emphasis on patient survival and cellular chemosensitivity. Acta Oncol 2012;51:69–76.
- Ghasimi S, Haapasalo H, Eray M, Korhonen K, Brännström T, Hedman H, et al. Immunohistochemical analysis of LRIG proteins in meningiomas: Correlation between estrogen receptor status and LRIG expression. J Neurooncol 2012;108:435–41.
- Rouam S, Moreau T, Broët P. Identifying common prognostic factors in genomic cancer studies: A novel index for censored outcomes. BMC Bioinformatics 2010;11:150.
- Li F, Yang W, Guo D, Hu Z, Xu H, Ye Z. LRIG1 combined with cisplatin enhances bladder cancer lesions via a novel pathway. Oncol Rep 2011;25:1629–37.
- Qi XC, Xie DJ, Yan QF, Wang YR, Zhu YX, Qian C, et al. LRIG1 dictates the chemo-sensitivity of temozolomide (TMZ) in U251 glioblastoma cells via down-regulation of EGFR/topoisomerase-2/Bcl-2. Biochem Biophys Res Commun 2013;437:565–72.
- Bai L, McEachern D, Yang CY, Lu J, Sun H, Wang S. LRIG1 modulates cancer cell sensitivity to Smac mimetics by regulating TNFα expression and receptor tyrosine kinase signaling. Cancer Res 2012;72:1229–38.

