Abstract
Human use of antibiotics has driven the selective enrichment of pathogenic bacteria resistant to clinically used drugs. Traditionally, the selection of resistance has been considered to occur mainly at high, therapeutic levels of antibiotics, but we are now beginning to understand better the importance of selection of resistance at low levels of antibiotics. The concentration of an antibiotic varies in different body compartments during treatment, and low concentrations of antibiotics are found in sewage water, soils, and many water environments due to natural production and contamination from human activities. Selection of resistance at non-lethal antibiotic concentrations (below the wild-type minimum inhibitory concentration) occurs due to differences in growth rate at the particular antibiotic concentration between cells with different tolerance levels to the antibiotic. The minimum selective concentration for a particular antibiotic is reached when its reducing effect on growth of the susceptible strain balances the reducing effect (fitness cost) of the resistance determinant in the resistant strain. Recent studies have shown that resistant bacteria can be selected at concentrations several hundred-fold below the lethal concentrations for susceptible cells. Resistant mutants selected at low antibiotic concentrations are generally more fit than those selected at high concentrations but can still be highly resistant. The characteristics of selection at low antibiotic concentrations, the potential clinical problems of this mode of selection, and potential solutions will be discussed.
Introduction
The use and over-use of antibiotics in human medicine, animal husbandry, and agriculture during the last 70 years have created a very strong selective pressure on bacteria, resulting in the emergence and spread of pathogenic bacteria resistant to every drug used. The resistance situation in some parts of the world is now becoming critical and has led to a serious lack of effective treatments for many bacterial infections and a fear that we are at the verge of a global post-antibiotic era (Citation1–3).
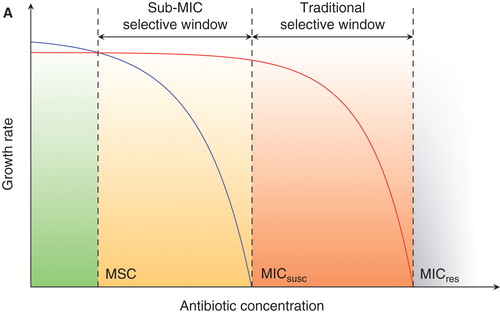
The underlying mechanisms of resistance development have been studied extensively in particular pathogen–antibiotic combinations. However, the broader questions of the driving forces behind where and how resistance arises and is selected and how resistance genes spread between different bacteria and different environments are complex and still not completely understood. Traditionally, selection of resistant bacteria has been suggested to occur at high antibiotic concentrations exceeding the minimal inhibitory concentration (MIC) of susceptible bacteria, since susceptible cells will no longer grow and are therefore outcompeted by resistant ones (Citation4). Recently, however, attention has been paid to how concentrations of antibiotics lower than the MIC of susceptible cells, sub-MIC levels, affect bacteria and what selective effects they have on emergence and enrichment of resistant bacteria. It is intuitive that for most kinds of antibiotics a bacterium will still experience a reduction in growth even at concentrations just below the MIC even if growth is not completely prevented (). A resistant bacterium will have a competitive advantage at all concentrations of an antibiotic at which the growth reduction of the susceptible strain is larger than the cost of resistance (orange + red areas in ). With experimental procedures that are sensitive enough to detect even very small differences in fitness we could therefore define a so-called minimum selective concentration (MSC) as the lowest concentration of an antibiotic that still selects for a given resistance mutation.
Figure 1. Schematic representation of growth rates as a function of antibiotic concentration. (MICsusc = minimal inhibitory concentration of the susceptible strain; MICres = minimal inhibitory concentration of the resistant strain; MSC = minimal selective concentration.) In green is the concentration range below the MSC in which the susceptible strain (blue line) will outcompete the resistant strain (red line) due to fitness cost of resistance. Orange (sub-MIC selective window) and red (traditional mutant selective window) indicate concentration intervals where the resistant strain will outcompete the susceptible strain due to the selective effect of antibiotic. Reproduced from (Citation5).
During antibiotic treatment of humans or animals the antibiotic concentration in the body is generally high, but there can be considerable variation both over the course of treatment and between different body compartments (Citation6–8). Depending on the specific nature of the antibiotic used (tissue distribution, clearance, etc.) the differences in concentration during treatment can result in low-level selection. In the outside environment, antibiotic concentrations due to natural production by micro-organisms and human contamination are typically much lower, in the range of μg/L to ng/L (Citation9–11). Bacteria are therefore exposed to low antibiotic concentrations in many different environments.
Sub-MIC selection
Two pioneering studies were recently published where slightly different approaches were used to measure the lowest concentration of different antibiotics that could selectively enrich for resistant bacteria (Citation5,12). Liu et al. determined the MSC for a ciprofloxacin-resistant Escherichia coli mutant (S83L) and an E. coli strain carrying tetracycline resistance on a Tn10 transposon to 1/5 and 1/20 of the MIC of the susceptible strain, respectively (Citation12). The assay used was based on competition at different antibiotic concentrations between a wild-type strain and a resistant strain differing only by the respective resistance marker and a lac-deletion for blue(wt)/white(resistant mutant) screening of colonies on plates to measure the ratio of the respective strain over time.
Gullberg et al. used a slightly different approach to determine MSC by competition between resistant and susceptible strains differentially expressing fluorescent proteins and determined the ratio between strains with very high resolution using flow cytometry (Citation5). There are two main advantages with this approach. First, there is a very small difference in fitness between the expression of the respective fluorescent proteins (YFP and CFP). This means that the effect on fitness of the resistance determinant is the main factor influencing the MSC. Second, a very high resolution is obtained by counting 100,000 cells per measurement. By such means differences in fitness as low as 0.2% can be detected (Citation5,13). With this approach the MSCs for three different antibiotics (ciprofloxacin, streptomycin, and tetracycline) were determined for a number of different resistant mutants in both E. coli and Salmonella Typhimurium LT2 (). The ciprofloxacin mutant (S83L) with a MSC 1/5 of the MICsusc in Liu et al. had a substantially lower MSC in Gullberg et al., 1/230-fold below the MICsusc (). Most likely, this difference is due to the very low difference in fitness between the wild type and the S83L mutant (0.2%) measured in Gullberg et al. (Citation5).
Table I. MSC for different resistance mutations. From (Citation5).
It is important to note that the MSC is intrinsically coupled to the fitness of the particular resistant mutant, since the cost of resistance has to be compensated for by the reduction in growth of the susceptible strain inferred by the antibiotic. This is most extensively shown by the S83L mutation. In comparison with other ciprofloxacin mutants in the same study it is evident that the fitness cost and not the actual resistance level of the mutation has the main effect on the MSC. For example, the resistance level of the D87N mutant is 5-fold higher than the efflux mutants (ΔacrR and ΔmarR). Still, they have the same MSC (). This means that in order to be selected at sub-MIC concentrations the mutant has to have a low fitness cost in contrast to when selected at high concentrations where a high MIC is more important.
The fact that antibiotic levels several hundred-fold below the MIC of the susceptible strains can select resistant bacteria means that the sub-MIC selective window is much larger than the traditional selective window. In effect this means that concentrations of antibiotics commonly found in sewage water in European countries and the USA (see (Citation9,14,15) and references therein) are high enough to enrich for resistant bacteria. Furthermore, the European Union has set the maximum allowed combined concentration of ciprofloxacin and enrofloxacin in milk for human consumption to be less than 100 ng/mL (Citation16), levels up to 1000-fold above the selective concentrations found by Gullberg et al. (Citation5).
De novo selection of resistance at sub-MIC concentrations
Gullberg et al. also showed that the initial ratio of resistant:susceptible bacteria did not have any effect on the MSC (Citation5). Ratios down to 1/10,000 resistant:susceptible bacteria still gave the same selective advantage at the tested antibiotic concentrations. This means that any spontaneous resistant mutant in a population will be selectively enriched at concentrations exceeding the MSC for that mutation. To test if sub-MIC concentrations could select for de novo resistant mutants Gullberg et al. also passaged wild-type Salmonella Typhimurium LT2 in liquid culture containing a concentration 1/4 of the MIC for streptomycin and E. coli in liquid culture containing 1/10 of the MIC for ciprofloxacin. Population analysis every 100 generations showed that resistant mutant populations arose rapidly and continued to increase in the population in both experiments. Interestingly, the resistance levels of the arising mutants were up to 32-fold above MICsusc for streptomycin and 8-fold above MICsusc for ciprofloxacin, 128-fold and 80-fold higher than the concentrations in which they were cycled. This means that the selection with sub-MIC concentrations of antibiotics not only enrich low-level resistance mutants but also those with clinically relevant levels of resistance.
Resistance mechanisms selected at sub-MIC
Since the fitness cost and not the level of resistance is the most influential parameter for selection of resistant cells at low levels of antibiotics, de novo selected mutants enriched at sub-MIC are expected to have very low fitness costs. It is therefore unlikely that the one-step high-level resistant mutants with relatively high fitness costs commonly found when high-level selection is performed will be selected (Citation17). Instead, accumulation of mutations giving increasing resistance levels but having very low fitness costs is predicted to occur. Such step-by-step evolution towards clinical resistance is well known from for example fluoroquinolone resistance development where no single mutation has been found to give clinical resistance by itself (Citation8,18,19). Possibly sub-MIC selection will also include previously unknown resistance mutations. Indeed, there are preliminary results from whole-genome sequencing of sub-MIC selected mutants confirming this (Citation5).
Problems with sub-MIC selection
There are several reasons why sub-MIC-selected resistant mutants are potentially more problematic than those selected at high concentrations (Citation20). First, since selection for high fitness is strong at sub-MIC levels of antibiotics it is less likely that the resulting resistance is reversed in the absence of antibiotic, either by mutation or by competition with more fit susceptible bacteria. It is therefore predicted that sub-MIC-selected resistant mutants will be more stable in bacterial populations than those selected by high concentrations (Citation21). Second, the rate with which resistance mutations will arise is expected to be higher at low concentrations of antibiotics. Non-lethal concentrations of antibiotics mean that the bacterial population is not eradicated as with high levels of drug where only pre-existing resistant mutants will survive. Instead, at sub-MIC the population will continue to grow allowing a larger effective population size and a continuing supply of possible resistance mutations. Also, many antibiotics (fluoroquinolones, aminoglycosides, beta-lactams) have been found to increase the rate of mutations at non-lethal concentrations thereby further adding to the possible supply of resistance mutations (Citation22–28). Third, low levels of antibiotics have been shown to increase homologous recombination rates, stimulate horizontal gene transfer, and activate integrating genetic elements (Citation29–34). This means that sub-lethal concentrations allow an increased supply of resistance mutations as well as increasing the genetic rearrangements that are involved in mobilization and spread of resistance. Fourth, the advantage of mutator phenotypes when accumulation of several mutations are needed for high-level resistance (Citation35–37) also means that sub-MIC selection is likely to enrich for mutators that will have a higher likelihood of generating multi-resistance.
Reducing the problem
What effect sub-MIC selection has on development of antibiotic resistance in clinically important bacteria is unknown and very hard to estimate. However, it is desirable to try to avoid such selection both during treatment of patients and even more so in animal husbandry (from growth-promoting use of antibiotics at non-therapeutic levels) and in environments downstream of the actual use of the antibiotics (water environments, sewage plants, and soils). Of course a general reduction in total antibiotic consumption will also reduce the presence of sub-MIC antibiotic levels, in particular in the environment. However, if sub-MIC selection in environmental settings is deemed to be a substantial way by which resistance determinants are selected and, more importantly, horizontally spread between different bacteria (e.g. from environmental bacteria to human pathogens), it is desirable also to implement methods by which we can break this chain of transmission. The two most straight-forward interventions are either to collect antibiotics at the source (e.g. by urine separation in hospitals or by limiting runoff from animal facilities) or to destroy drugs downstream at the sewage plants. Oxidative destruction (mainly by ozone treatment) of many different pharmaceuticals (not only antibiotics) has been discussed and tested (Citation38,39). This would also reduce the impact on the environment of drugs such as contraceptives, anti-depressants, pain-killers, etc. that severely affect water-dwelling organisms. An additional advantage of oxidative treatment is that pathogens (resistant or not) such as bacteria, viruses, and parasites would also be killed, further limiting their possibility to make their way back to the human population.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- van Dyck Hoffmann A. editor. Action plan against the rising threats from antimicrobial resistance. Communication from the Commission to the European Parliament and the Council; 2011. pp 1–17; Report No.: COM (2011) 748. Available at: http://ec.europa.eu/health/antimicrobial_resistance/policy/index_en.htm.
- Laxminarayan R, Duse A, Wattal C, Zaidi AKM, Wertheim HFL, Sumpradit N, et al. Antibiotic resistance—the need for global solutions. Lancet Infect Dis. 2013;13:1057–98.
- Antibiotic Resistance Threats in the United States, 2013. CDC. USA: U. S. Department of Health and Human Services, Centers for Disease Control and Prevention; 2013. pp 1–114. Available at: http://www.cdc.gov/drugresistance/threat-report-2013/.
- Drlica K. The mutant selection window and antimicrobial resistance. J Antimicrob Chemother. 2003;52:11–17.
- Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, et al. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 2011;7:e1002158.
- Baquero F, Negri MC, Morosini MI, Blazquez J. Antibiotic-selective environments. Clin Infect Dis. 1998;27:S5–11.
- Baquero F, Negri MA-C. Challenges: selective compartments for resistant microorganisms in antibiotic gradients. Bioessays. 1997;19:731–6.
- Baquero F, Negri MC, Morosini MI, Blázquez J. The antibiotic selective process: concentration-specific amplification of low-level resistant populations. Ciba Found Symp. 1997;207:93–105; discussion 105–11.
- Kümmerer K. Antibiotics in the aquatic environment – A review – Part I. Chemosphere. 2009;75:417–34.
- Thiele-Bruhn S. Pharmaceutical antibiotic compounds in soils – a review. J Plant Nutr Soil Sci. 2003;166:145–67.
- Chander Y, Kumar K, Goyal SM, Gupta SC. Antibacterial activity of soil-bound antibiotics. J Environ Qual. 2005;34:1952.
- Liu A, Fong A, Becket E, Yuan J, Tamae C, Medrano L, et al. Selective advantage of resistant strains at trace levels of antibiotics: a simple and ultrasensitive color test for detection of antibiotics and genotoxic agents. Antimicrob Agents Chemother. 2011;55:1204–10.
- Lind PA, Tobin C, Berg OG, Kurland CG, Andersson DI. Compensatory gene amplification restores fitness after inter-species gene replacements. Mol Microbiol. 2010;75:1078–89.
- Segura PA, François M, Gagnon C, Sauvé S. Review of the occurrence of anti-infectives in contaminated wastewaters and natural and drinking waters. Environ Health Perspect. 2009;117:675–84.
- Wahlberg C, Björlenius B, Paxéus N. Fluxes of 13 selected pharmaceuticals in the water cycle of Stockholm, Sweden. Water Sci Technol. 2011;63:1772–80.
- Ashwin H, Stead S, Caldow M, Sharman M, Stark J, de Rijk A, et al. A rapid microbial inhibition-based screening strategy for fluoroquinolone and quinolone residues in foods of animal origin. Anal Chim Acta. 2009;637:241–6.
- Hughes D, Andersson DI. Selection of resistance at lethal and non-lethal antibiotic concentrations. Curr Opin Microbiol. 2012;15:555–60.
- Baquero F. Low-level antibacterial resistance: a gateway to clinical resistance. Drug Resist Updat. 2001;4:93–105.
- Orlen H, Hughes D. Weak mutators can drive the evolution of fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 2006;50:3454–6.
- Andersson DI, Hughes D. Evolution of antibiotic resistance at non-lethal drug concentrations. Drug Resist Updat. 2012;15:162–72.
- Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–71.
- Ysern P, Clerch B, Castańo M, Gibert I, Barbé J, Llagostera M. Induction of SOS genes in Escherichia coli and mutagenesis in Salmonella typhimurium by fluoroquinolones. Mutagenesis. 1990;5:63–6.
- Ren L, Rahman MS, Humayun MZ. Escherichia coli cells exposed to streptomycin display a mutator phenotype. J Bacteriol. 1999;181:1043–4.
- Miller C. SOS Response induction by β-lactams and bacterial defense against antibiotic lethality. Science. 2004;305:1629–31.
- Balashov S, Humayun MZ. Mistranslation induced by streptomycin provokes a RecABC/RuvABC-dependent mutator phenotype in Escherichia coli cells. J Mol Biol. 2002;315:513–27.
- Perez-Capilla T, Baquero MR, Gomez-Gomez JM, Ionel A, Martin S, Blazquez J. SOS-independent induction of dinB transcription by β-lactam-mediated inhibition of cell wall synthesis in Escherichia coli. J Bacteriol. 2005;187:1515–18.
- Baharoglu Z, Mazel D. Vibrio cholerae triggers SOS and mutagenesis in response to a wide range of antibiotics: a route towards multiresistance. Antimicrob Agents Chemother. 2011;55:2438–41.
- Thi TD, López E, Rodríguez-Rojas A, Rodríguez-Beltrán J, Couce A, Guelfo JR, et al. Effect of recA inactivation on mutagenesis of Escherichia coli exposed to sublethal concentrations of antimicrobials. J Antimicrob Chemother. 2011;66:531–8.
- Barr V, Barr K, Millar MR, Lacey RW. β-Lactam antibiotics increase the frequency of plasmid transfer in Staphylococcus aureus. J Antimicrob Chemother. 1986;17:409–13.
- Doucet-Populaire F, Trieu-Cuot P, Dosbaa I, Andremont A, Courvalin P. Inducible transfer of conjugative transposon Tn1545 from Enterococcus faecalis to Listeria monocytogenes in the digestive tracts of gnotobiotic mice. Antimicrob Agents Chemother. 1991;35:185–7.
- Bahl MI, Sørensen SJ, Hansen LH, Licht TR. Effect of tetracycline on transfer and establishment of the tetracycline-inducible conjugative transposon Tn916 in the guts of gnotobiotic rats. Appl Environ Microbiol. 2004;70:758–64.
- López E, Elez M, Matic I, Blázquez J. Antibiotic-mediated recombination: ciprofloxacin stimulates SOS-independent recombination of divergent sequences in Escherichia coli. Mol Microbiol. 2007;64:83–93.
- Couce A, Blázquez J. Side effects of antibiotics on genetic variability. FEMS Microbiol Rev. 2009;33:531–8.
- Cantón R, Morosini MI. Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol Rev. 2011;35:977–91.
- Mao EF, Lane L, Lee J, Miller JH. Proliferation of mutators in A cell population. J Bacteriol. 1997;179:417–22.
- Taddei F, Godelle B, Radman M, Maynard-Smith J, Toupance B, Gouyon PH. Nature. 1997;387:700–2.
- Shaver AC, Dombrowski PG, Sweeney JY, Treis T, Zappala RM, Sniegowski PD. Fitness evolution and the rise of mutator alleles in experimental Escherichia coli populations. Genetics. 2002;162:557–66.
- Esplugas S, Bila DM, Krause LGT, Dezotti M. Ozonation and advanced oxidation technologies to remove endocrine disrupting chemicals (EDCs) and pharmaceuticals and personal care products (PPCPs) in water effluents. J Hazard Mater. 2007;149:631–42.
- Wahlberg C, Björlenius B, et al. Fluxes of 13 selected pharmaceuticals in the water cycle of Stockholm, Sweden. Water Science and Technology: A Journal of the International Association on Water Pollution Research. 2011;63:1772–80.

