Abstract
Calcific aortic valve disease (CAVD) is a progressive pathological condition with no effective pharmacological therapy. To identify novel molecular pathways as potential targets for pharmacotherapy, we studied microRNA (miRNA) profiles of heavily stenotic aortic valves (AS). One of the most upregulated miRNAs in AS valves compared to control valves was miR-125b (1.4-fold; P < 0.05). To identify CAVD-related changes in gene expression, DNA microarray analysis was performed, including an intermediate fibro(sclero)tic stage of the disease. This revealed changes especially in genes related to inflammation and immune response, including chemokine (C-C motif) ligand 3 (CCL3) and 4 (CCL4). CCL3 mRNA level was increased 3.9-fold (P < 0.05) when AS valves were compared to control valves, and a 2.5-fold increase (P < 0.05) in CCL4 gene expression was observed when fibro(sclero)tic valves were compared to control valves. Both CCL3 and CCL4 localized to macrophages by immunofluorescence. To identify chemokine–miRNA target pairs, data from miRNA target prediction databases were combined with valvular miRNA and mRNA expression profiles. MiR-125b was computationally predicted to target CCL4, as confirmed experimentally in cultured human THP-1 macrophages. Collectively, miR-125b and CCL4 appear to be involved in the progression of CAVD and may offer novel therapeutic and diagnostic strategies related to this disease.
Calcific aortic valve disease is characterized by changes in microRNA and inflammatory gene expression.
Both chemokine CCL4 and miR-125b are upregulated in CAVD, and miR-125b is a potential regulator of CCL4 in cultured macrophages.
Introduction
Calcific aortic valve disease (CAVD) is the most common form of valvular heart disease in the Western world. Milder degrees of aortic valve calcification, i.e. aortic sclerosis, can be seen in 26% of people aged > 65 years, whereas severe calcification with impaired valve leaflet motion (termed aortic stenosis, AS) has a prevalence of 2% (Citation1). CAVD leads to a significantly increased risk of mortality; currently the only treatment for symptomatic CAVD is surgical or transcatheter aortic valve replacement (Citation2). Given that the majority of patients are elderly and currently the only effective treatment to CAVD is valve replacement, there is an increasing demand for a pharmacologic intervention.
Calcification of the aortic valve was formerly considered to be an age-related degenerative process, but emerging evidence has revealed it as an actively regulated disease (Citation3) and the outcome of several overlapping processes, including calcification, ossification, neovascularization, and inflammation (Citation3). These processes are also typical determinants of atherosclerosis, but despite pathophysiological similarities only about 50% of patients with CAVD have clinically significant atherosclerosis (Citation4). Although some progress has been made in understanding the factors driving CAVD, the molecular pathogenesis and signaling mechanisms underlying the progression of AS are poorly understood.
MicroRNAs are short, non-coding RNA molecules that generally function as negative regulators of gene expression by complementary binding of 3’-UTRs of messenger RNAs (mRNAs). This can lead to either blockade of protein translation or the complete degradation of the miRNA–mRNA complex. It has been estimated that miRNAs may regulate up to one-third of the human genome (Citation5). The involvement of miRNAs in cardiovascular diseases ranges from primary risk factors to pathophysiological processes (Citation6). However, the involvement of miRNAs in the pathogenesis of AS remains to be uncovered.
In this study we compared miRNA profiles of stenotic and control aortic valves to determine the miRNAs involved in the development of CAVD. To identify the possible miRNA targets, a genome-wide gene expression analysis of aortic valves was performed. This DNA microarray analysis revealed changes in valvular gene expression associated especially with inflammation, chemokines and their receptors being the dominant group of genes upregulated in calcific aortic valves. Next, to identify miRNA target pairs with confidence, we combined data from miRNA target prediction databases and expression profiles of miRNA and mRNA focusing on inflammatory response. We found that miR-125b was upregulated in stenotic valves, and it was found to target chemokine (C-C motif) ligand 4 (CCL4) in silico; this relationship was further confirmed in vitro in cultured human THP-1 macrophages.
Materials and methods
An expanded Methods section is available in the supplementary material online: patient demographics (Supplementary Table 1, to be found online at http://informahealthcare.com/doi/abs/10.3109/07853890.2015.1059955), analysis of miRNA, miRNA array, DNA microarray, pathway analysis, analysis of RNA, Bioinformatic microRNA target prediction, miRNA transfection, ELISA, histological and immunohistological analysis, and statistical analysis.
Table I. The 10 most up- and downregulated genes in stenotic valves compared to control valves.
Results
Valvular calcification and neovascularization are markedly elevated in patients with aortic stenosis.
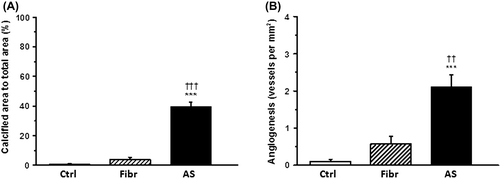
All the valves in the AS group were markedly calcified, the proportion of calcified area to the total aortic valve area being 39.6% ± 3.1% (), while only weak calcification was observed in the fibro(sclero)sis group. There were no macroscopic signs of calcification in the control group. Factor VIII immunostaining was used to visualize the density of neovascularization in aortic valves. Neo-angiogenesis was significantly activated in the AS group in comparison to other groups, while only few vascular structures were found in control and fibro(sclero)tic valves ().
Figure 1. Valvular calcification and neovascularization in aortic valves. A: The proportion of calcified area in human aortic valve cusps to total aortic valve cusp area. B: Number of vascular structures per square millimeter in human aortic valve cusps. Results are mean ± SEM. ***P < 0.001 versus control group (Ctrl; n = 13); †††P < 0.001, ††P < 0.01 versus fibro(sclero)sis group (Fibr; n = 12). Aortic stenosis group (AS; n = 28).
Differentially expressed miRNAs in CAVD
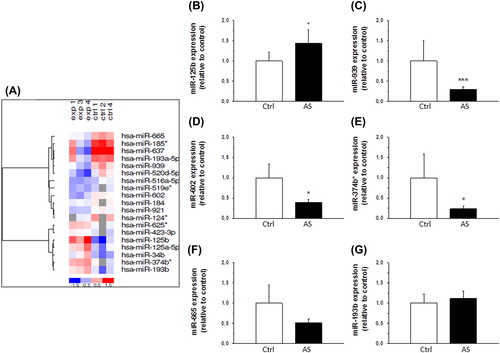
MiRCURYTM LNA array microRNA expression profiling (Exiqon, Vedbaek, Denmark) by using commercial miRCURYTM LNA Arrays (v.10.0) containing 1154 probes of miRBase was conducted to determine differentially expressed miRNA molecules in stenotic aortic valves when compared to control valves (n = 3 in both groups). The analysis showed a total of 19 miRNAs to be significantly altered (P < 0.05) as shown in the heat map in . The expression levels of the most significantly altered miRNAs (-125b, -939, -602, -665, -374b*, and -193b) were further validated by quantitative RT-PCR in another larger sample set (Ctrl, n = 6; AS, n = 20) (). The differential expression of miRNAs -125b, -939, -602, and -374b* was statistically significant (P < 0.05).
Figure 2. Expression of miRNAs in human aortic valves. A: A heat map diagram showing the results of two-way hierarchical clustering of miRNAs. Each row represents a miRNA and each column represents a sample. The miRNA clustering tree is shown on the left. The color scale shown at the bottom illustrates the relative expression level of miRNA across the samples: red color, expression level above mean; blue color, expression lower than the mean; gray color, signal below background. B–G: Expression levels of miR-125b (B), miR-939 (C), miR-602 (D), miR-374b* (E), miR-665 (F), and miR-193b (G) in human aortic valve cusps in control (Ctrl, n = 6) and aortic stenosis group (AS, n = 20) as measured by quantitative RT-PCR. Results are expressed as ratios of miRNA to SNORA73 (mean ± SEM). *P < 0.05, ***P < 0.001 versus control group (Student's t test or Mann–Whitney U test).
DNA microarray analysis of micro-RNA targets genes in CAVD
To identify potential miRNA target genes, we determined differentially expressed genes during aortic valve calcification by evaluating gene expression profiles of control, fibro(sclero)tic, and stenotic valves (n = 5 in each group). Selected DNA microarray results were confirmed by comparison with mRNA levels obtained by quantitative RT-PCR; both methods showed changes of equal magnitude (Supplementary Table 4, to be found online at http://informahealthcare.com/doi/abs/10.3109/07853890.2015.1059955).
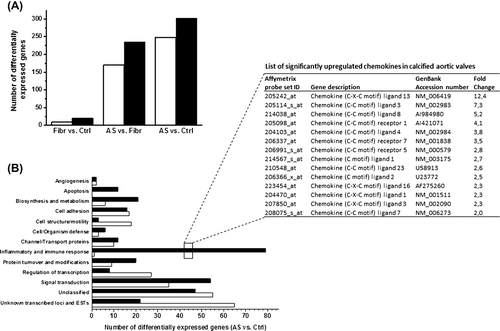
Our analysis identified 302 up- and 248 downregulated transcripts (fold-change ≥ 2-fold) when the AS group was compared to the control group (; and Supplementary Table 5, to be found online at http://informahealthcare.com/doi/abs/10.3109/07853890.2015.1059955). The majority of upregulated genes were genes related to inflammation and immune response, as well as to signal transduction. In contrast, the majority of downregulated genes with known function were signal transduction proteins and regulators of transcription (). The top 10 most up- and downregulated genes in stenotic versus control valves are shown in . Several chemokines were among the top upregulated genes during development of AS (). Importantly, the majority of changes in gene expression was seen at the late phase of the disease: 235 genes were upregulated and 170 genes downregulated more than 2-fold when the AS group was compared with the fibro(sclero)tic group (; and Supplementary Figure 1A, to be found online at http://informahealthcare.com/doi/abs/10.3109/07853890.2015.1059955), whereas altogether only 34 genes showed differential expression at the early phase of the disease (fibro(sclero)tic versus control group) (; and Supplementary Figure 1B, to be found online at http://informahealthcare.com/doi/abs/10.3109/07853890.2015.1059955). The differentially expressed genes during development of AS are shown in Supplementary Tables 5–7 (to be found online at http://informahealthcare.com/doi/abs/10.3109/07853890.2015.1059955).
Figure 3. DNA microarray analysis of calcific aortic valve disease. A: The open bars show the number of downregulated and the solid bars upregulated genes in human aortic valve cusps in fibro(sclero)sis group (Fibr) versus control group (Ctrl), aortic stenosis group (AS) versus Fibr, and AS versus Ctrl. B: Functional classification of differentially expressed genes in human aortic valve cusps in AS versus Ctrl group revealing upregulation of many chemokines (listed and ranked by fold change in the inset table).
The complete list of significantly up- or downregulated genes in stenotic versus control valves was loaded into the STRING database to identify potential interactions between differentially expressed genes. Of those genes, 103 had confirmed regulatory connections with biological evidence in databases (list of genes in Supplementary Table 8, to be found online at http://informahealthcare.com/doi/abs/10.3109/07853890.2015.1059955). The main interaction network of CAVD is shown in Supplementary Figure 2 (to be found online at http://informahealthcare.com/doi/abs/10.3109/07853890.2015.1059955). Central signaling molecules in this pathway are chemokines and their receptors, including C-X-C chemokine ligand 3 (CXCL3), C-C chemokine receptor 5 (CCR5), and T-cell antigen receptor CD247, implicating the importance of inflammatory and T-cell responses in CAVD.
MiRNA-125b modulates chemokine CCL4
To identify miRNA target pairs with confidence, we combined data from miRNA target prediction databases and expression profiles of both miRNA and mRNA with focus on inflammatory response. DNA microarray analysis showed that especially many chemokines such as CCL3 and CCL4 were significantly upregulated in CAVD. Interestingly, one of the most prominent dysregulated miRNAs was miR-125b, which has previously been linked to regulation of inflammatory genes in vascular smooth muscle cells (VSMCs) of diabetic mice (Citation7).
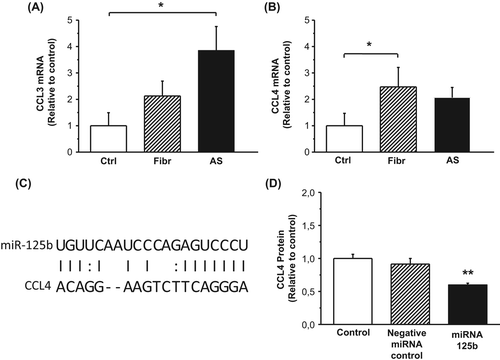
Quantitative RT-PCR analysis confirmed that CCL3 and CCL4 mRNA levels were significantly increased in the different stages of CAVD ( and B). According to MicroCosm targets, CCL4 may be regulated by miR-125b (). To test this hypothesis, we transfected human THP-1 macrophages with a mimic for miR-125b and measured the corresponding mRNA and protein levels of CCL4. As shown in , a statistically significant decrease in CCL4 protein secretion was seen in response to miRNA-125b transfection as measured by ELISA, which corresponded to changes in the CCL4 mRNA levels (Supplementary Figure 3, to be found online at http://informahealthcare.com/doi/abs/10.3109/07853890.2015.1059955).
Figure 4. A–B: Gene expression of chemokine (C-C motif) ligand 3 (CCL3) (A) and CCL4 (B) in control valves and in the different stages of calcific aortic valve disease. Results are expressed as ratios of respective mRNA to 18S (mean ± SEM). *P < 0.05 versus control group. Ctrl, n = 9; Fibr, n = 7; AS, n = 25. C: Correlation of CCL4 mRNA 3’-UTR seed region and miR-125b sequence indicating a potential regulatory relationship, as predicted by MicroCosm software. D: Production of CCL4 protein by human THP-1 macrophages 24 hours after transfection with transfection agent alone (mock), scrambled miRNA (negative miRNA control), or miRNA-125b. Results from three independent experiments are presented as mean ± SEM.
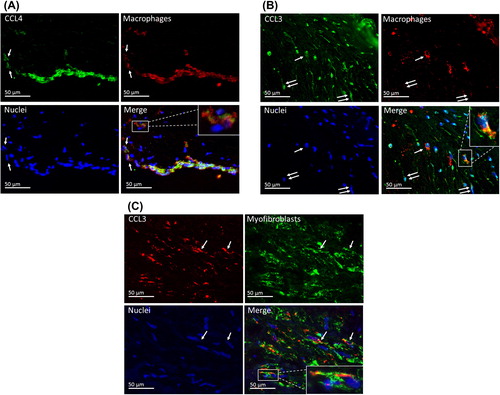
Localization of CCL3 and CCL4 in aortic valve cusps was evaluated by immunofluorescence revealing that CCL4 is expressed in macrophages (). CCL3 is expressed in some macrophages as well as myofibroblasts ( and C). These results were verified using immunohistochemistry (Supplementary Figure 5, to be found online at http://informahealthcare.com/doi/abs/10.3109/07853890.2015.1059955).
Figure 5. Chemokine localization in aortic valves. A: CCL4 is expressed in macrophages. In this area, there is a strand of subendothelial macrophages as well as some scattered macrophages (arrows), in which the co-localization of CCL4 and CD68 is seen. Detailed image of a single macrophage is shown in the inset. B: CCL3 is expressed in some macrophages (arrow) and CD68-negative spindle cells (double arrows). Detailed image of a single macrophage is shown in the inset. C: In this focus, there are several α-SMA-positive myofibroblasts. Some of these co-express CCL3 (arrows).
Discussion
CAVD is recognized as a complex, actively regulated biological process including inflammation, lipid retention, and calcification (Citation3). Therefore, a better understanding of the molecular mechanisms driving CAVD pathogenesis in aortic valves is needed to provide new targets for novel therapeutic interventions. In this study we performed miRNA profiling of normal and calcified aortic valves to determine involvement of miRNAs in the pathogenesis of CAVD. We characterized targets of these miRNAs by conducting genome-wide gene expression profiling of different stages of CAVD. The most prominent changes were observed in genes involved in inflammatory and immune response including chemokines CCL3 and CCL4. We identified the miRNAs regulating these inflammatory genes by combining data from miRNA target prediction databases and expression profiles of both miRNA and mRNA. We found that miR-125b targeted CCL4, and a miR-125b mimic decreased expression of CCL4 also in THP-1 macrophages. Altogether, our results suggest that miR-125b and CCL4 are associated with the progression of CAVD.
As a major finding, we detected a significant downregulation of miRNAs -939, -602, and -374b* as well as increased expression of miR-125b in calcified aortic valves. In further analysis we focused on miR-125b, previously linked to inflammation (Citation7) and calcification (Citation8), the two key processes of CAVD. Other miRNAs identified by our array may also have important regulatory potential. For example, miR-939 was recently shown to regulate nitric oxide synthase, an important factor in endothelial function (Citation9). MiR-374b may be a regulator of CCTTA enhancer binding protein-β (C/EBP-β) (Citation10), a key factor in macrophage polarization (Citation11). Also, miR-602 could be a regulator of Ras association domain-containing protein 1 (RASSF1) (Citation12), which has been further shown to regulate apoptosis (Citation13). These findings suggest that miR-dysregulation during CAVD may have multifactorial impact on the underlying pathological processes.
In search of possible miRNA targets, our genome-wide gene expression analysis demonstrated that the majority of upregulated genes were involved in activation of inflammatory and immune response in each phase of the disease process, chemokines and immunoglobulins being the largest functional families. This supports previous studies emphasizing the importance of chronic inflammation in AS. For example, inflammatory infiltrates within stenotic valve leaflets have been associated with an active remodeling process (Citation14). Our pathway analysis further highlighted the biological relevance of inflammatory response in CAVD since chemokines and their receptors as well as mediators of the T-cell response such as CD247 were central signaling molecules in this network. To our knowledge, there is only one earlier large-scale gene expression study of CAVD (Citation15). Intriguingly, matrix metallopeptidase 12, CXCL5, tryptophan 2,3-dioxygenase, and collagen XIα1 were among the top 10 upregulated genes in both the earlier and our study. We also identified other well-known regulators of CAVD including cathepsin S (Citation16), tryptase (Citation17), as well as osteopontin, bone sialoprotein II, and osteoprotegerin (Citation18), in agreement with the established role of extracellular matrix modulation in the pathophysiology of CAVD (Citation15). Of note, in the study of Bosse et al. (Citation15) only heavily stenotic and control valves were compared, while in our study an intermediate fibro(sclero)tic group enabled studying earlier stages of CAVD. Therefore, the important finding of our study is that there were only minor changes at the gene expression level in the early phase of the disease. This implies that the irreversible pathological changes occur in the later stages of the disease progression, underscoring the importance of early intervention and prevention of CAVD.
Most of the genes we uncovered have not previously been known to be associated with CAVD, although they are linked to the key processes of atherosclerosis, e.g. angiopoietin-1 to angiogenesis (Citation19) and BCL2-related protein A1 to apoptosis (Citation20). In addition, we identified several transcription factors, including runt-related transcription factor 3 (Runx3), activation transcription factor-3 (ATF-3), and interferon regulatory factor 8 (IRF-8), which could be of importance for the development of CAVD. Runx3 is an osteogenic gene regulating e.g. expression of osteopontin (Citation21), ATF-3 has been linked to macrophage activation (Citation22), and IRF-8 is a transcription factor that affects inflammatory cell populations in atherosclerosis (Citation23). Clearly, further studies are needed to clarify their role in pathogenesis of CAVD.
In this study we detected an increased expression of the inflammatory chemokines CCL3 and CCL4. Although the expression of several chemokines such as CCL18 (Citation24) and CCL23 (Citation25) has been linked to the development of atherosclerotic lesions in humans, the role of chemokines in CAVD is largely unknown and limited to expression studies (Citation15,Citation26). In addition, there is evidence that CCL4 could also function as a heterodimer with CCL3 (Citation27), which was also upregulated in stenotic valves. Similarly, the expression of ATF-3 and CCR5 was upregulated in our array. ATF-3 has been shown to regulate CCL4 expression (Citation28), and CCR5 is also a receptor for CCL4 (Citation29). Our immunofluorescence studies of CCL3 and CCL4 support the evidence that these chemokines localize in macrophage-rich microenvironments, possibly being secreted by the macrophages in order to attract more inflammatory cells into the inflamed valve (Citation30). We also demonstrate that CCL3 is in part expressed by myofibroblasts as a part of the inflammatory response. Interestingly, it has recently been shown that modulation of the chemokine system may be an effective way to reduce atherosclerosis and inflammation (Citation31–33), suggesting that pharmacological modulation of the chemokine system could be an effective treatment strategy in the development of CAVD.
We discovered that CCL4 could be a potential target for miR-125b regulation, since the transfection of miR-125b into THP-1 macrophages significantly reduced both CCL4 mRNA and protein levels. Interestingly, we discovered a concomitant increase of expression of both miR-125b and CCL4 in the valves, which could be due to the overall inflammatory condition in the valvular tissue. In VSMCs, miR-125b has been shown to regulate inflammatory genes (Citation7) as well as vascular calcification (Citation8). Therefore, it is likely that the tissue-level increase of miR-125b is related to the inflammatory and osteogenic response of valvular cells, although it is a negative regulator of CCL4 in macrophages. Consequently, the upregulation of miR-125b may occur in cells other than macrophages. It has also been shown that under certain circumstances miRNAs may also induce the upregulation of target genes (Citation34). This would provide an alternative explanation for the simultaneous upregulation of both miR-125b and CCL4. It is also very likely that CCL4 is regulated by a network of several factors which remain beyond the scope of this study. Further research is needed to determine the mechanism of this miRNA-mediated regulation in CAVD.
Study limitations
In addition to the chemokine regulation presented here, the effects of miRNAs in aortic valve surely extend to hundreds of genes. Mapping these interactions further will provide a more comprehensive understanding of CAVD progression. To our knowledge, the only previous study on miRNAs in aortic valve leaflets was limited to a small population of congenitally bicuspid aortic valves, and the authors were unable to detect any genes targeted by the identified miRNAs (Citation35). It should also be noted that there was some heterogeneity between patient groups, including age. Nevertheless, for both miRNA and mRNA arrays, the valves were matched for age, sex, and treatments. Finally, the miRNA-targeted transcription was studied only in THP-1 macrophages. Based on our immunofluorescence-based localization studies, macrophages were the likely source of CCL4, although we cannot rule out the contribution of other cell types at tissue level.
Conclusions
In human stenotic aortic valves miR-125b is upregulated, whereas miR-939, miR-602, and miR-374b* are downregulated, revealing that miRNAs are associated with the progression of CAVD. We showed that chemokines are among the most upregulated inflammatory genes in CAVD, and experimental evidence demonstrated that miR-125b is one of the regulators of CCL4 in THP-1 macrophages. Altogether, our data suggest a novel miRNA-mediated regulatory mechanism in CAVD and may offer novel therapeutic and diagnostic strategies related to this disease.
Supplementary material available online
Supplementary Methods and Supplementary Figures 1–6 to be found online at http://informahealthcare.com/doi/abs/10.3109/07853890.2015.1059955
iann_a_1059955_sm0738.pdf
Download PDF (705.6 KB)Acknowledgements
We thank Dr Jussi Vuoristo for help with microarrays, and Kati Viitala, Manu Tuovinen, and Mari Jokinen for expert technical assistance.
Funding:
This study was supported by Academy of Finland Center of Excellence Funding (H.R. grant no. 266661) and the grants 276747 and 284504 (J.R.), the Finnish Foundation for Cardiovascular Research (J.R., H.R., P.O.), Paavo Nurmi Foundation, Ida Montin Foundation (P.O.), Orion Research Foundation (P.O.), Aarne Koskelo Foundation (P.O.), and Sigrid Jusélius Foundation (H.R.). The Wihuri Research Institute is maintained by the Jenny and Antti Wihuri Foundation.
Declaration of interest:
The authors report no conflicts of interest.
References
- Stewart BF, Siscovick D, Lind BK, Gardin JM, Gottdiener JS, Smith VE, et al. Clinical factors associated with calcific aortic valve disease. Cardiovascular Health Study. J Am Coll Cardiol. 1997;29:630–4.
- Rodes-Cabau J. Transcatheter aortic valve implantation: current and future approaches. Nat Rev Cardiol. 2011;9:15–29.
- Helske S, Kupari M, Lindstedt KA, Kovanen PT. Aortic valve stenosis: an active atheroinflammatory process. Curr Opin Lipidol. 2007;18: 483–91.
- Qian J, Chen Z, Ge J, Ma J, Chang S, Fan B, et al. Relationship between aortic valve calcification and the severity of coronary atherosclerotic disease. J Heart Valve Dis. 2010;19:466–70.
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, et al. Systematic discovery of regulatory motifs in human promoters and 3’ UTRs by comparison of several mammals. Nature. 2005;434:338–45.
- Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013;123:11–18.
- Villeneuve LM, Kato M, Reddy MA, Wang M, Lanting L, Natarajan R. Enhanced levels of microRNA-125b in vascular smooth muscle cells of diabetic db/db mice lead to increased inflammatory gene expression by targeting the histone methyltransferase Suv39h1. Diabetes. 2010;59: 2904–15.
- Goettsch C, Rauner M, Pacyna N, Hempel U, Bornstein SR, Hofbauer LC. miR-125b regulates calcification of vascular smooth muscle cells. Am J Pathol. 2011;179:1594–600.
- Guo Z, Shao L, Zheng L, Du Q, Li P, John B, et al. miRNA-939 regulates human inducible nitric oxide synthase posttranscriptional gene expression in human hepatocytes. Proc Natl Acad Sci U S A. 2012;109:5826–31.
- Pan S, Zheng Y, Zhao R, Yang X. MicroRNA-130b and microRNA-374b mediate the effect of maternal dietary protein on offspring lipid metabolism in Meishan pigs. Br J Nutr. 2013;109:1731–8.
- Ruffell D, Mourkioti F, Gambardella A, Kirstetter P, Lopez RG, Rosenthal N, et al. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc Natl Acad Sci U S A. 2009;106:17475–80.
- Yang L, Ma Z, Wang D, Zhao W, Chen L, Wang G. MicroRNA-602 regulating tumor suppressive gene RASSF1A is overexpressed in hepatitis B virus-infected liver and hepatocellular carcinoma. Cancer Biol Ther. 2010;9:803–8.
- Oh HJ, Lee KK, Song SJ, Jin MS, Song MS, Lee JH, et al. Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res. 2006;66:2562–9.
- Cote N, Mahmut A, Bosse Y, Couture C, Pagé S, Trahan S, et al. Inflammation is associated with the remodeling of calcific aortic valve disease. Inflammation. 2013;36:573–81.
- Bosse Y, Miqdad A, Fournier D, Pepin A, Pibarot P, Mathieu P. Refining molecular pathways leading to calcific aortic valve stenosis by studying gene expression profile of normal and calcified stenotic human aortic valves. Circ Cardiovasc Genet. 2009;2:489–98.
- Helske S, Syvaranta S, Lindstedt KA, Lappalainen J, Oörni K, Mäyränpää MI, et al. Increased expression of elastolytic cathepsins S, K, and V and their inhibitor cystatin C in stenotic aortic valves. Arterioscler Thromb Vasc Biol. 2006;26:1791–8.
- Helske S, Lindstedt KA, Laine M, Mäyränpää M, Werkkala K, Lommi J, et al. Induction of local angiotensin II-producing systems in stenotic aortic valves. J Am Coll Cardiol. 2004;44:1859–66.
- Pohjolainen V, Taskinen P, Soini Y, Rysä J, Ilves M, Juvonen T, et al. Noncollagenous bone matrix proteins as a part of calcific aortic valve disease regulation. Hum Pathol. 2008;39:1695–701.
- Fukuhara S, Sako K, Noda K, Zhang J, Minami M, Mochizuki N. Angiopoietin-1/Tie2 receptor signaling in vascular quiescence and angiogenesis. Histol Histopathol. 2010;25:387–96.
- Bae J, Hsu SY, Leo CP, Zell K, Hsueh AJ. Underphosphorylated BAD interacts with diverse antiapoptotic Bcl-2 family proteins to regulate apoptosis. Apoptosis. 2001;6:319–30.
- Cheng HC, Liu YP, Shan YS, Huang CY, Lin FC, Lin LC, et al. Loss of RUNX3 increases osteopontin expression and promotes cell migration in gastric cancer. Carcinogenesis. 2013;34:2452–9.
- Xu YZ, Thuraisingam T, Marino R, Radzioch D. Recruitment of SWI/SNF complex is required for transcriptional activation of the SLC11A1 gene during macrophage differentiation of HL-60 cells. J Biol Chem. 2011;286:12839–49.
- Doring Y, Soehnlein O, Drechsler M, Shagdarsuren E, Chaudhari SM, Meiler S, et al. Hematopoietic interferon regulatory factor 8-deficiency accelerates atherosclerosis in mice. Arterioscler Thromb Vasc Biol. 2012;32:1613–23.
- Hagg DA, Olson FJ, Kjelldahl J, Jernås M, Thelle DS, Carlsson LM, et al. Expression of chemokine (C-C motif) ligand 18 in human macrophages and atherosclerotic plaques. Atherosclerosis. 2009;204:e15–20.
- Kim CS, Kang JH, Cho HR, Blankenship TN, Erickson KL, Kawada T, et al. Potential involvement of CCL23 in atherosclerotic lesion formation/progression by the enhancement of chemotaxis, adhesion molecule expression, and MMP-2 release from monocytes. Inflamm Res. 2011;60:889–95.
- Dorfmuller P, Bazin D, Aubert S, Weil R, Brisset F, Daudon M, et al. Crystalline ultrastructures, inflammatory elements, and neoangiogenesis are present in inconspicuous aortic valve tissue. Cardiol Res Pract. 2010;2010:685926.
- Guan E, Wang J, Norcross MA. Identification of human macrophage inflammatory proteins 1alpha and 1beta as a native secreted heterodimer. J Biol Chem. 2001;15:12404–9
- Chandrasekar B, Deobagkar-Lele M, Victor ES, Nandi D. Regulation of chemokines, CCL3 and CCL4, by interferon gamma and nitric oxide synthase 2 in mouse macrophages and during Salmonella enterica serovar typhimurium infection. J Infect Dis. 2013;207:1556–68.
- Samson M, Labbe O, Mollereau C, Vassart G, Parmentier M. Molecular cloning and functional expression of a new human CC-chemokine receptor gene. Biochemistry. 1996;35:3362–7.
- Weber C, Schober A, Zernecke A. Chemokines: key regulators of mononuclear cell recruitment in atherosclerotic vascular disease. Arterioscler Thromb Vasc Biol. 2004;24:1997–2008.
- Cipriani S, Francisci D, Mencarelli A, Renga B, Schiaroli E, D’Amore C, et al. Efficacy of the CCR5 antagonist maraviroc in reducing early, ritonavir-induced atherogenesis and advanced plaque progression in mice. Circulation. 2013;127:2114–24.
- Leuschner F, Dutta P, Gorbatov R, Novobrantseva TI, Donahoe JS, Courties G, et al. Therapeutic siRNA silencing in inflammatory monocytes in mice. Nat Biotechnol. 2011;29:1005–10.
- de Jager SC, Bot I, Kraaijeveld AO, Korporaal SJ, Bot M, van Santbrink PJ, et al. Leukocyte-specific CCL3 deficiency inhibits atherosclerotic lesion development by affecting neutrophil accumulation. Arterioscler Thromb Vasc Biol. 2013;33:e75–83.
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4.
- Nigam V, Sievers HH, Jensen BC, Sier HA, Simpson PC, Srivastava D, et al. Altered microRNAs in bicuspid aortic valve: a comparison between stenotic and insufficient valves. J Heart Valve Dis. 2010;19:459–65.

