Abstract
Objective. Evidence exists that arterial stiffness, i.e. an independent predictor of cardiovascular and all-causes mortality, has a genetic component. The 9p21 region is associated with a greater susceptibility to coronary disease. Whether this can be ascribed to the fact that genes located on chromosome 9p may also regulate arterial stiffness is largely unknown, however. We evaluate the influence of single nucleotide polymorphisms (SNPs) from 9p on carotid–femoral pulse wave velocity (C-F PWV), measured via the Complior method, in a cohort of 821 hypertensive subjects. Design. The selected tagSNPs were screened with a custom-designed 384-plex VeraCode GoldenGate Genotyping assay on Illumina BeadXpress Reader platform. Association analysis was done using PLINK considering C-F PWV as a quantitative trait (linear regression assuming an additive model) adjusting for sex, age, systolic blood pressure and body mass index (BMI). We used false discovery rate (FDR) to account for multiple testing. Results. Although none of the 384 SNPs was significant after adjusting for multiple testing, probably due to the small sample size of the study population, a trend of association with C-F PWV was observed for rs300622 and rs2381640. Conclusions. These data suggest that SNPs located on chromosome 9p may affect arterial stiffness. Further studies are needed to confirm our finding on a larger sample and define the physiopathological link of the present results.
Introduction
Arterial stiffness is well known to be an independent predictor of cardiovascular and all-causes mortality (Citation1), especially when it is measured as carotid–femoral pulse wave velocity (C-F PWV). Moreover, it has been demonstrated an association between the degree of arterial stiffening and the entity of coronary vessels disease (Citation2).
The arterial stiffening process is associated with aging, hypertension, diabetes mellitus and chronic kidney diseases (Citation3,Citation4); nevertheless, a consistent fraction of arterial stiffness variability still remains largely unexplained. Arterial stiffness heritability is estimated to be between 0.13 and 0.54 (Citation5,Citation6), suggesting that arterial stiffness is influenced by genetic status. Candidate genes studies have shown a potential role of genetic polymorphisms located in renin– angiotensin–aldosterone system, NO synthase, G proteins, Elastin, type 1 Collagen, metallopeptidase 3 and 9 genes (Citation7–10). Recently, genome-wide association studies (GWAS) have shown an association between measures of arterial stiffness and loci located in different chromosomes (chromosomes 2, 7, 13, 15), while their physiopathology link with arterial stiffening process remains largely unclear (Citation11–13). In 2007, McPherson et al. (Citation14) identified a 58-kilobase interval on chromosome 9p21 that has been associated with coronary heart disease in six independent samples (more than 23,000 participants) from four Caucasian populations. Since then, the 9p21 region was confirmed to be associated with the development of coronary heart disease in many studies, and was also shown to be associated with intracranial and abdominal aneurisms. However, to date, there are few studies that evaluated the association between genes located in this region and cardiovascular intermediate phenotypes (Citation15,Citation16).
In the present paper, we examined the effects of single nucleotide polymorphisms (SNPs) in the short arm of chromosome 9 on arterial stiffness. We assessed arterial stiffness with the C-F PWV method, in accurately phenotyped hypertensive individuals.
Participants and methods
Ethical considerations
The protocol of this study has been approved by institutional ethics review committees at the relevant organizations involved. The study protocol complies with the Declaration of Helsinki (as revised in 2004) (World Medical Association Declaration of Helsinki, Ethical principles for Medical Research Involving Human Subjects. http://www.wma.net/e/policy/b3.htm) All participants provided informed written consent.
Essential hypertensive cohort
Eight hundred and twenty-seven hypertensive outpatients, aged 18–80 years old, followed by the Hypertension Centre of S. Gerardo Hospital, Monza, Italy, were enrolled from September 2006 until October 2008.
Exclusion criteria were the presence of secondary hypertension, chronic renal disease, chronic pulmonary disease, substance abuse and history of cancer.
A mercury sphygmomanometer was used to measure clinic BP twice with the patient in the sitting position for at least 5 min and with the arm placed at heart level. The first and fifth phase of Korotkoff sounds were taken as systolic blood pressure (SBP) and diastolic blood pressure (DBP) values while the cuff was deflated at a rate of 2 mmHg/s. We then assessed BP twice with a semiautomatic device. The average of the two manual and two semiautomatic BP readings was recorded. Hypertension was defined as SBP ≥ 140 mmHg and DBP ≥ 90 mmHg, or as the reported use of antihypertensive drugs. Levels of fasting serum glucose, serum total cholesterol, high-density lipoprotein (HDL), low-density lipoprotein (LDL) cholesterol and serum triglycerides were determined from venous blood sample.
Arterial stiffness was evaluated by pulse wave velocity (PWV) between the carotid and the femoral artery of the same side with the patient in the supine position (Citation17). The pressure pulse waveforms were simultaneously obtained from the two arterial sites using an automatic device (Complior System, Colson, Les Lilas, Francia) and their distance calculated by taking the distance between hip and neck via a rigid paediatric tape. Two measurements were obtained in each patient and the average value was used for the analysis.
Single nucleotide polymorphism selection and genotyping
We selected 384 tagSNPs on the short arm of chromosome 9 from HapMap Phase II (http://www.hapmap.org) using the linkage disequilibrium (LD) tagSNP selection approach (Citation18). Selection criteria included: SNPs with validation data, successful predictive genotyping scores for Illumina GoldenGate assays, a minor allele frequency (MAF)≥ 0.05 and a pairwise LD threshold of r2 ≥ 0.80 for Caucasians. A further refinement was made selecting SNPs belonging to coding, intronic and 5′ and 3′ untranslated regions with similar proportions. We also included 7 SNPs belonging to the matrix metallopeptidase 9 (MMP9), elastin (ELN) and endothelin receptor type A (ET-A) genes and to the cyclin-dependent kinase inhibitor 2A (CDKN2A) and cyclin-dependent kinase inhibitor 2B (CDKN2B) regions, which were previously been shown to be associated with arterial stiffness (Citation15,Citation16).
Genomic DNA was extracted from 300 μl of fresh peripheral blood using Wizard Genomic DNA Purification kit (Promega, Madison, WI, USA) and then resuspended in 70 μl of pure water. DNA aliquots were stocked at − 80°C. Genotyping was performed on the on Illumina BeadXpress Reader platform (Illumina Inc., San Diego, CA, USA). Genotypes were called using GenomeStudio. To validate Illumina results, 100 samples were screened further for six of the 384 SNPs on the ABI Prism 3130 Avant Automatic Sequencer (Applied Biosystems).
Statistical analysis
Quality checking included manual review of all cluster plots, genotype frequency, call rate and deviation from Hardy–Weinberg equilibrium. For Hardy–Weinberg equilibrium, a p-value threshold of 5 × 10− 4 was used. Analysis of variance and the χ2 test were used to compare continuous and categorical variables, respectively, between gender. Association analysis was done using PLINK (Citation19). We performed linear regression using the additive model and adjusting for covariates determined by stepwise regression. The covariates used for multivariate adjustments were age, sex, SBP and body mass index (BMI), which are known to be major determinant of the arterial stiffening process. Because testing multiple SNPs could lead to false-positive associations, we applied the false discovery rate (FDR) (Citation20).
Results
As shown in , 57% of the total cohort was represented by males, they were younger than females, had higher BMI, DBP, total cholesterol, triglycerides, glucose, creatinine and left ventricular mass index (LVMI). They had significantly lower heart rate (cardiac frequency electrocardiogram, CF ECG), HDL- and LDL-cholesterol. Eighteen per cent of men and 16% of females were current smokers, and 81% men and 78% females were on anti-hypertensive treatment. Thirty-one of the 384 autosomal SNPs selected, after quality control, were excluded because of poor clustering, two were excluded because of MAF less than 0.01, three were excluded because of deviation from Hardy–Weinberg equilibrium and 348 SNPs were included in the final analysis. Genotyping was successful in 99.5% of individuals.
Table I. Demographic characteristics of the cohort overall and by gender.
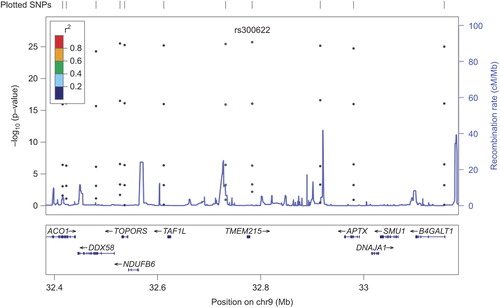
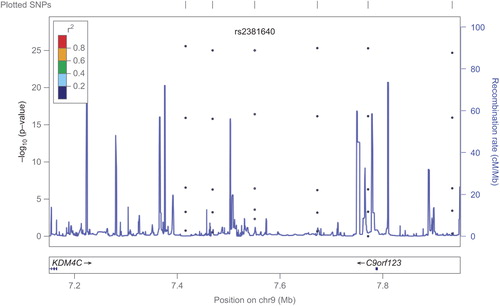
Single marker association analysis showed nominal significance for rs300622 (beta = 0.017, SE = 0.006, p-value = 0.003) and rs2381640 (beta = 0.013, SE = 0.004, p-value = 0.003) as shown in and in the regional association plots ( and ). The allelic distribution of the two SNPs is also reported in . The major allele for rs300622 is A (frequency = 0.87) while the minor is C (frequency = 0.13); the major allele for rs2381640 is C (frequency = 0.7) and the minor is T (frequency = 0.3). SNP rs300622 is located in flanking 3′ untranslated region of the TMEM215 gene on the short arm of chromosome 9 (9p21.1) Rs2381640 is located in flanking 5 untranslated region of the TRIM5 pseudogene (positive strand) and on the RPL4P5 pseudogene (negative strand) of chromosome 9p24.1. These two SNPs lost statistical significance after multiple testing correction.
Table II. Allelic distribution of rs300622 and rs2381640.
Discussion
In an accurately phenotyped cohort of essential hypertensives from Northern Italy, we identified two SNPs on the short arm of chromosome 9 that showed an association trend with C-F PWV.
Rs300622 is located in 9p21.1 on the TMEM215 gene, which encodes for the transmembrane protein 215. Rs2381640 is located in 9p24.1 on the TRIM5 pseudogene (positive strand) and on the RPL4P5 pseudogene (negative strand). TRIM5 encodes for a structural protein while RPL4P5 encodes for a ribosomal protein. The TMEM215 gene and transmembrane protein 215 have been studied in host genetic contribution to vaccine response study (Citation21). The TRIM family proteins share a conserved arrangement of three adjacent domains, which constitutes the tripartite-motif for which the family is named; the TRIM5alpha has a C-terminal B30.2/SPRY domain, which is the major determinant of viral target specificity (Citation22). The RPL4P5 pseudogene function has been evaluated in a study on protein–protein interaction, which suggests the importance of pseudogene in the understanding of protein function and cellular processes (Citation23). Thus, a biologically plausible association of genetic results with the atherosclerotic process is not self-evident and remains unclear; further studies are needed to better dissect the mechanistic basis of these results.
The results of our study deserve some further comments. First, while not surviving to the multiple analysis correction, the results of the present study suggest a role of the short arm of chromosome 9 on arterial stiffness. Since arterial stiffness is one of the most powerful predictors of coronary heart disease (Citation1), one can extrapolate that both the genetic basis of coronary artery disease and arterial stiffness are on the short arm of chromosome 9. An alternative possible interpretation is that, as arterial stiffness is one of the major determinants of the vascular atherogenic process, this physiopathological link might be genetically related. Another observation refers to the pulse wave velocity measurement itself. Indeed, in this study, the measurement was obtained in all enrolled patients, with no exception, and this supports the idea of its feasibility as an alternative, easy and chip target organ damage parameter in hypertension.
Our study has some strengths as well as limitations. The strengths refer to the fact that our discovery cohort is a sample of accurately phenotyped hypertensive patients. This allowed us to account for several potential confounders like SBP, age, gender and BMI. Furthermore, in our study, arterial stiffness was measured as C-F PWV, which is regarded as the gold standard method with a strong prognostic value. Our study also has several limitations, however. First, the association results disappear after multiple testing correction. In addition, we were not able to look up our nominally significant results in an independent sample, as rarely do investigators include pulse wave velocity analysis when they phenotype essential hypertensive patients and we could not find a cohort with the same measurement in the same conditions. It is thus possible that with a larger cohort and a replicate, the association we are suggesting in this paper can be confirmed. Moreover, we were not able to replicate our result in normotensive healthy individuals and this will be the objective of future research programmes on the issue.
Conclusions
In conclusion, the 9p region may influence arterial stiffness and the identification of genes that influence intermediate cardiovascular phenotypes may improve early diagnosis and preventive measures. Further studies are necessary to confirm the suggestions and define the physiopathological link of the present results.
Declaration of interest: The paper has been made possible by the following grant: the EU-MASCARA project has received funding from the European Community's Seventh Framework Programme (FP7/2007–2013) under Grant Agreement no 278249. The “NEDD” project, accepted for a grant by Regione Lombardia in 2010.
References
- Vlachopoulos C, Aznaouridis K, Stefanadis C. Prediction of cardiovascular events and all-cause mortality with arterial stiffness: A systematic review and meta-analysis. J Am Coll Cardiol. 2010;55:1318–1327.
- Weber T, Auer J, O’Rourke MF. Increased arterial wave reflections predict severe cardiovascular events in patients undergoing percutaneous coronary interventions. Eur Heart J. 2005; 26:2657–2663.
- McEniery CM,Yasmin, Hall IR. Normal vascular aging: Differential effects on wave reflection and aortic pulse wave velocity: The Anglo-Cardiff Collaborative Trial (ACCT). J Am Coll Cardiol. 2005;46:1753–1760.
- Giannattasio C, Failla M, Capra A. Increased arterial stiffness in normoglycemic normotensive offspring of type 2 diabetic parents. Hypertension. 2008;51:182–187.
- Sayed-Tabatabaei FA, van Rijn MJ, Schut AF. Heritability of the function and structure of the arterial wall: Findings of the Erasmus Rucphen Family (ERF) study. Stroke. 2005;36: 2351–2356.
- Mitchell GF, DeStefano AL, Larson MG. Heritability and a genome-wide linkage scan for arterial stiffness, wave reflection, and mean arterial pressure: The Framingham Heart Study. Circulation. 2005;112:194–199.
- Wojciechowska W, Staessen JA, Stolarz K. Association of peripheral and central arterial wave reflections with the CYP11B2 -344C allele and sodium excretion. J Hypertens. 2004;22:2311–239.
- Nürnberger J, Opazo Saez A, Mitchell A. The T-allele of the C825T polymorphism is associated with higher arterial stiffness in young healthy males. J Hum Hypertens. 2004;18: 267–271.
- Medley TL, Kingwell BA, Gatzka CD. Matrix metalloproteinase-3 genotype contributes to age-related aortic stiffening through modulation of gene and protein expression. Circ Res. 2003;92:1254–1261. Epub 2003 May 15.
- Medley TL, Cole TJ, Dart AM. Matrix metalloproteinase-9 genotype influences large artery stiffness through effects on aortic gene and protein expression. Arterioscler Thromb Vasc Biol. 2004;24:1479–1484. Epub 2004 Jun 10.
- DeStefano AL, Larson MG, Mitchell GF. Genome-wide scan for pulse pressure in the National Heart, Lung and Blood Institute's Framingham Heart Study. Hypertension. 2004;44:152–155. Epub 2004 Jun 21.
- Björck HM, Länne T, Alehagen U. Association of genetic variation on chromosome 9p21.3and arterial stiffness. J Intern Med. 2009;265:373–381. Epub 2008 Oct 25.
- Tarasov KV, Sanna S, Scuteri A. COL4A1 is associated with arterial stiffness by genome-wide association scan. Circ Cardiovasc Genet. 2009;2:151–158. Epub 2009 Feb 18.
- McPherson R, Pertsemlidis A, Kavaslar N. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491.
- Yasmin, McEniery CM, O’Shaughnessy KM. Variation in the human matrix metalloproteinase-9 gene is associated with arterial stiffness in healthy individuals. Arterioscler Thromb Vasc Biol. 2006;26:1799–1805.
- Iwai N, Kajimoto K, Kokubo Y, Tomoike H. Extensive genetic analysis of 10 candidate genes for hypertension in Japanese. Hypertension. 2006;48:901–907.
- Laurent S, Cockcroft J, Van Bortel. European Network for Non-invasive Investigation of Large Arteries. Expert consensus document on arterial stiffness: Methodological issues and clinical applications.Eur Heart J. 2006;27:2588–2605.
- Purcell S, Neale B, Todd-Brown K. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575.
- Benjamini Y HY. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Stat Soc Ser B. 1995;57:289–300.
- Pajewski NM, Shrestha S, Quinn CP. A genome-wide association study of host genetic determinants of the antibody response to anthrax vaccine adsorbed. Vaccine. 2012;30: 4778–4784.
- Newman RM, Hall L, Kirmaier A. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008 Feb 29;4(2):e1000003.
- Stelzl U, Worm U, Lalowski M. A human protein–protein interaction network: A resource for annotating the proteome. Cell. 2005;123:957–968.
- Levy D, Larson MG, Benjamin EJ. Framingham Heart Study 100K Project: Genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007;8:S3.