Abstract
DNA repair gene polymorphisms may affect DNA repair capacity and modulate susceptibility to end-stage renal disease (ESRD). We aimed to determine the association of polymorphisms in xeroderma pigmentosum complementation group D (XPD) and X-ray cross-complementing group 1 (XRCC1) with ESRD development. Polymorphisms in XPD codons 312 and 751 and XRCC1 codon 399 were genotyped in 98 patients undergoing hemodialysis and 102 healthy controls using polymerase chain reaction and restriction fragment length polymorphism. Patients having XRCC1-399 Arg/Gln genotype or XRCC1-399 Gln/Gln genotype had a significantly higher risk of ESRD than those with XRCC1-399 Arg/Arg [odds ratio (OR): 2.48; 95% confidence intervals (CI): 1.36–4.52; p = 0.004 and OR: 4.05; 95% CI: 1.19–13.73; p = 0.03, respectively]. We also found a significantly higher frequency of the XRCC1 399Gln allele in patients with ESRD than in controls (OR: 2.22; 95% CI: 1.16–4.25; p = 0.02). Combination of the Arg/Gln or Gln/Gln genotypes of XRCC1 Arg399Gln polymorphism with Asp/Asn or Asn/Asn genotypes of XPDAsp312Asn or with the Lys/Gln or Gln/Gln genotypes of XPD Lys751Gln was significantly associated with the development of ESRD. Haplotypes association showed that association of Gln allele of XRCC1 Arg399Gln polymorphism with the Asn allele of XPDAsp312Asn polymorphism (p = 0.004) or Gln allele of XRCC1 Arg399Gln polymorphism with the Gln allele of XPD Lys751Gln polymorphism (p = 0.003) was highly significantly associated with the development of ESRD. This study revealed that XRCC1 Arg399Gln polymorphism may confer increased risk for the development of ESRD. Furthermore, larger studies should be conducted to confirm these results.
Introduction
Worldwide, the increasing number of individuals in end-stage renal disease (ESRD) represents a serious health, social and economic problem. The causes of ESRD are heterogeneous, ranging from infectious diseases and metabolic multisystemic disease to congenital and genetic disorders. This variety of possible etiologies makes it difficult to identify the mechanisms involved in its pathogenesis. In many cases, the etiology remains unknown.Citation1 Traditional risk factors such as age, black race, male gender, smoking, hypertension, hyperlipidemia, diabetes and obesity cannot explain the complete individual susceptibility to development of ESRD and accompanying complications that essentially determine the clinical outcome. Because of clinically experienced inter-individual differences, an important role of genetic predisposition might be assumed.
Patients with ESRD display enhanced genomic damage.Citation2 If genomic damage is left unrepaired or is repaired with errors, mutations of critical genes may occur and may result in an enhanced cancer risk. Genomic damage may also be involved in initiation as well as progression of cardiovascular diseases.Citation3 ESRD is associated with a high incidence of cancers and cardiovascular diseases.Citation4 Among the several pathogenic mechanisms suggested to explain these phenomena, there are uremia per se, micro inflammation and oxidative stress,Citation5 which involves the whole cell structure (proteins, membrane lipids, carbohydrates and DNA).Citation6 Oxidative stress is enhanced in patients with ESRD.Citation7 It has been reported that oxidative stress can induce DNA damage, such as base modifications and strand breaks.Citation8 DNA repair enzymes continuously monitor chromosomes to correct damaged nucleotide residues generated by exposure to cytotoxic compounds or carcinogens. Repair of oxidative DNA damage is mediated by both base excision repair (BER) and nucleotide excision repair (NER) mechanisms. It has been hypothesized in many studies that polymorphisms in DNA repair genes reduce their capacity to repair DNA damage and thereby lead to increased cancer or other disease susceptibility.Citation9
Xeroderma pigmentosum complementation group D (XPD) encodes a helicase, which participates in both NER and basal transcription as part of the transcription factor IIH (TFIIH). As XPD is important in multiple cellular tasks and rare XPD mutations result in genetic diseases, XPD polymorphisms may operate as genetic susceptibility factors. Several single nucleotide polymorphisms (SNPs) in XPD gene exons have been identified; of them, Asp312Asn and Lys751Gln polymorphisms are the most common.Citation10 XPD Asp312Asn in exon 10 causes an amino acid substitution in a conserved region of XPD. XPD Lys751Gln in exon 23 also causes an amino acid substitution in the C-terminal part of the protein.Citation10,Citation11 These polymorphisms may produce the most relevant change in XPD function and affect different protein interactions, diminish the activity of TFIIH complexes, influence DNA repair capacity and alter the genetic susceptibility for diseases.Citation11,Citation12 SNPs of XPD gene have been studied in relation to lung cancerCitation13 and colorectal cancer,Citation14,Citation15 cataract and glaucoma development;Citation16,Citation17 also, it has been investigated in different hematological malignancies, such as acute myeloid and lymphoblastic leukemia.Citation18–20
X-ray cross-complementing group 1 (XRCC1), a DNA repair protein involved in single-strand breaks and BER pathway, has been reported to be responsible for the efficient repair of DNA damage caused by active oxygen, ionization and alkylating agents.Citation21 It is a multidomain protein that interacts with the nicked DNA and participates with at least three different enzymes, poly-ADP-ribose polymerase, DNA ligase III and DNA polymerase β, to repair single-strand breaks.Citation21 Three coding polymorphisms were identified in the XRCC1 gene at the codons 194 (Arg to Trp), 280 (Arg to His) and 399 (Arg to Gln).Citation10 Although the functional effects of these polymorphisms in XRCC1 have not been well known, amino acid changes at evolutionary conserved regions may alter its function. The XRCC1 polymorphisms have been extensively studied in relation to acute myeloid leukemia,Citation22,Citation23 acute lymphoblastic leukemia,Citation24,Citation25 chronic lymphocytic leukemia,Citation26 lymphoma,Citation27,Citation28 chronic myeloid leukemiaCitation29 and cardiovascular disease.Citation30 XRCC1 and XPD polymorphisms are also associated with increased risk of hepatocellular carcinoma.Citation31,Citation32
In this study, we aimed to examine the possible relationship between DNA repair enzymes (XPD Asp312Asn and XPD Lys751Gln and XRCC1 Arg399Gln) polymorphisms and the risk of developing ESRD in a sample from Egyptian cohort.
Methods
Study subjects
Ninety-eight patients with ESRD undergoing hemodialysis were enrolled in this controlled study (48 females and 50 males, mean age 47.8 ± 14.2 years, range: 20–80, median 50). All subjects were of Egyptian nationality. Time on dialysis of patients was 3.9 ± 3.7 years. Control group was formed by 102 healthy individuals (46 females and 56 males, mean age 46.3 ± 13.2 years, range: 21–76, median 48). The causes of ESRD were hypertension in 44 patients, diabetes mellitus (DM) in 11 patients, preeclampsia in 4 patients, drug induced in 3 patients, glomerulonephritis in 6 patients, obstructive uropathy in 5 patients, atrophic kidney in 3 patients, systemic lupus erythematosus in 5 patients, polycystic kidney in 2 patients, combined polycystic kidney and hypertension in 1 patient, combined DM and hypertension in 6 patients, amyloidosis and hypertension in 1 patient and unknown cause in 7 patients.
This study was approved by the ethical committee of the Faculty of Medicine, Menoufia University. All patients provided signed informed consent to provide a blood sample and to review the medical record for research purposes.
Genotyping
DNA was isolated from peripheral leucocytes using a GeneJET whole blood Genomic DNA Purification Kit (Thermo Scientific, Waltham, MA) according to the manufacturer’s instructions.
XPD genotypes were detected using a polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP) method. An Asp → Asn in exon 10 (codon 312) and a Lys → Gln in exon 23 (codon 751) were amplified to form an undigested fragments of 751 and 436 bp, respectively, using primers described by Batar et al.Citation33: 5′-CTG TTG GTG GGT GCC CGT ATC TGT TGG TCT-3′ (forward) and 5′-TAA TAT CGG GGC TCA CCC TGC AGC ACT TCC T-3′ (reverse) for codon 312; and 5′-GCC CGC TCT GGA TTA TAC G-3′ (forward) and 5′-CTA TCA TCT CCT GGC CCC C-3′ (reverse) for codon 751. PCR conditions were 94 °C for 3 min, followed by 38 cycles of 94°C for 45 s, 60 °C for 45 s, 72 °C for 60 s and a final extension step at 72 °C for 7 min. PCR products were digested with StyI (for codon 312) and PstI (for codon 751) (Thermo Scientific) at 37 °C overnight and analyzed on 3% agarose gels. StyI digestion resulted in two fragments of 507 and 244 bp for the wild-type homozygous (Asp/Asp); three fragments of 474, 244, and 33 bp for the variant homozygous (Asn/Asn); and four fragments of 507, 474, 244 and 33 bp for the heterozygous (Asp/Asn) (). PstI digestion resulted in two fragments of 290 and 146 bp for the wild-type homozygous (Lys/Lys); three fragments of 227, 146 and 63 bp for the variant homozygous (Gln/Gln); and four fragments at 290, 227, 146 and 63 bp for the heterozygous (Lys/Gln) ().
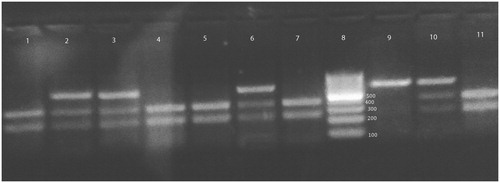
Figure 1. XPD 312 genotypes on ethidium bromide stained 3% agarose gel. Lanes 1, 2 and 7 are homozygous Asp/Asp genotype. Lanes 3, 4, 5 and 9 are heterozygous Asp/Asn genotype. Lane 6 is homozygous Asn/Asn genotype. Lane 8 is 100 bp ladder.
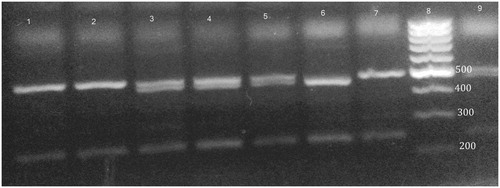
Figure 2. XPD 751 genotypes on ethidium bromide stained 3% agarose gel. Lanes 1, 6 and 9 are homozygous Lys/Lys genotype. Lanes 2 and 4 are heterozygous Lys/Gln genotype. Lanes 3, 5, 7 and 10 are homozygous Gln/Gln genotype. Lane 8 is 100 bp ladder.
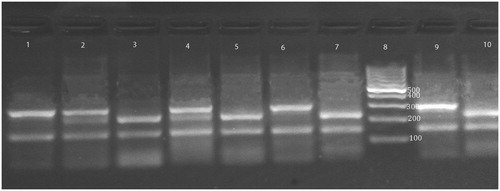
XRCC1 codon 399 genotype was detected using a PCR–RFLP method. An Arg → Gln in exon 10 (codon 399) were amplified to form an undigested fragments of 615 bp, using primers described by Batar et al.Citation33: 5′-TTG TGC TTT CTC TGT GTC CA-3′ (forward) and 5′-TCC TCC AGC CTT TTC TGA TA-3′ (reverse). PCR conditions were 94 °C for 3 min, followed by 30 cycles of 94 °C for 30 s, 56 °C for 1 min, 72 °C for 45 s and a final extension step at 72 °C for 10 min. The 615 bp PCR products were digested with MspI (Thermo Scientific) at 37 °C overnight and analyzed on 3% agarose gel. MspI digestion resulted in two fragments of 374 and 221 bp for wild-type homozygous (Arg/Arg); one fragment of 615 bp for variant homozygous (Gln/Gln); and three fragments of 615, 374 and 221 bp for variant heterozygous (Arg/Gln) ().
Statistical analysis
Data were processed using the Statistical Package for Social Science version 16 (SPSS, Chicago, IL). The ages of the patients and the controls were compared by the Student t test. The chi-square test was used to compare the gender distribution and to test the association between the genotypes and alleles in relation to the cases and controls. Deviation of genotype distribution from the Hardy–Weinberg equilibrium and haplotype association analysis were carried out using the web tool SNPStats. The odds ratios (ORs) and their 95% confidence intervals (95% CIs) were calculated to estimate the strength of the association between polymorphism genotype alleles of patients and controls. A value of p < 0.05 was considered statistically significant.
Results
The distributions of the XPDAsp312Asn, XPD-Lys751Gln and XRCC1-Arg399Gln genotypes did not display significant deviation from the Hardy–Weinberg equilibrium between patients and healthy individuals. There were no significant age and sex differences between patients and controls (p = 0.4).
There were a significant difference between frequencies for XRCC1-399 Arg/Gln genotype and XRCC1-399 Gln/Gln genotype in patients and controls (p = 0.004 and p = 0.03, respectively). We found that patients with XRCC1-399 Arg/Gln (OR: 2.48; 95% CI: 1.36–4.52) or XRCC1-399 Gln/Gln (OR: 4.05; 95% CI: 1.19–13.73) genotype had a significantly higher risk of ESRD than those with XRCC1-399 Arg/Arg genotype (). The Gln allele of this polymorphism was associated with increased risk of ESRD (OR: 2.22; 95% CI: 1.16–4.25; p = 0.02).
Table 1. Distribution of XPD and XRCC1 genotype polymorphisms in patients and controls.
No statistically significant differences were observed in the alleles or in the genotype frequencies of the XPD-Asp312Asn and XPD-Lys751Gln gene polymorphisms between the control group and the patients group ().
Regarding the effect of combined polymorphisms of XPD Asp312Asn, XPD Lys751Gln and XRCC1 Arg399Gln on the risk of ESRD development, the wild type genotypes for each gene were taken as references (). The analysis showed that combination of Arg/Gln or Gln/Gln genotypes of XRCC1 Arg399Gln polymorphism with Asp/Asn or Asn/Asn of XPDAsp312Asn polymorphism was significantly associated with the development of ESRD (p = 0.05). Furthermore, combination of Arg/Gln or Gln/Gln genotypes of XRCC1 Arg399Gln polymorphism with Lys/Gln or Gln/Gln genotypes of XPD Lys751Gln polymorphism was found to increase the risk of ESRD development (p = 0.01). However, no significant association was found between other compound polymorphisms and the risk of developing ESRD.
Table 2. Distribution of combined XPD and XRCC1 genotypes among ESRD patients and control.
The haplotype association analysis showed that the association of Gln allele of XRCC1 Arg399Gln polymorphism with the Asn allele of XPDAsp312Asn polymorphism (p = 0.004, OR: 8.35; 95% CI: 1.94–35.85; ) or Gln allele of XRCC1 Arg399Gln polymorphism with the Gln allele of XPD Lys751Gln polymorphism (p = 0.003, OR: 9.22; 95% CI: 2.14–39.71; ) was highly significantly associated with the development of ESRD. Furthermore, haplotypes association of the three codons revealed that association of Gln allele of XRCC1 399 with the Asn allele of XPD 312 and Gln allele of XPD 751 (p < 0.0001) was significantly associated with the development of ESRD ().
Table 3. Haplotype association of XPD 312 and XRCC1 399 with ESRD.
Table 4. Haplotype association of XPD 751 and XRCC1 399 with ESRD.
Table 5. Haplotype association of XPD 312, XPD 751 and XRCC1 399 with ESRD.
Discussion
Various studies have shown the existence of a large interindividual variation in repair of DNA damage induced by endogenous and exogenous insults and the individuals with less dramatic reduction in the capacity to repair DNA are observed at polymorphic frequency.Citation34 Such individuals with repair capacity below the population mean can be at increased risk of developing several chronic diseases. Data regarding the relationship between DNA repair enzymes XRCC1 and XPD polymorphisms and ESRD are limited.
In our study, all the examined subjects were genotyped for two repair genes, XRCC1 and XPD in order to analyze the possible influence of the genetically determined variations on susceptibility to ESRD.
Our study provides no evidence of a role of XPDAsp312Asn and Lys751Gln polymorphisms, both homozygous variant (Asn/Asn and Gln/Gln) and combined heterozygous + homozygous variant genotypes (Asp/Asn-Asn/Asn and Lys/Gln-Gln/Gln) in susceptibility to ESRD. We also found no significant association between the XPD 312Asn and 751Gln alleles and ESRD risk. Our findings are in agreement with the results reported by Trabulus et al.Citation35 in Turkish population. The frequencies of XPD 312Asn and 751Gln alleles in our ESRD patients were 0.35 and 0.37, whereas in controls they were 0.36 and 0.37, respectively. Trabulus et al.Citation35 analyzed the XPDAsp312Asn and Lys751Gln polymorphism on a cohort which included 136 dialysis patients and 147 controls from Turkey. The frequency of the variant XPD 312Asn and 751Gln alleles were 0.46 and 0.43 in cases and 0.40 and 0.43 in controls.
There may be some explanations regarding the results indicating no relationship between the polymorphisms of XPDAsp312Asn and Lys751Gln and the risk of ESRD in this study. First, the exposure and interaction of other genes participating in DNA damage recognition, repair and cell cycle regulation may have altered the effect of XPD and XRCC1 polymorphisms.Citation36 Second, ethnic, genetic and environmental differences in allele frequency for the investigated polymorphisms might also affect the results in genetic studies. Third, different levels of exposure of certain oxidative stimuli in different individuals may have also contributed to the association between the polymorphisms of the DNA repair genes and the risk of diseases. Fourth, DNA repair capacity among individuals is variable and it is genetically determined. Everyone has a unique combination of polymorphic traits that modify susceptibility and response to drugs, exogenous and endogenous chemical toxins and carcinogenic exposures.
Regarding XRCC1 codon 399, our results revealed a positive association between the XRCC1 399 Gln variant both homozygous (Gln/Gln) and combined heterozygous + homozygous variant genotypes (Arg/Gln + Gln/Gln) and the risk of ESRD. We also observed an association between variant XRCC1 399 Gln allele and the risk of ESRD. These results suggest that the XRCC1 Arg399Gln polymorphism may contribute to ESRD development. Our findings are in agreement with the study conducted by Trabulus et al.Citation35 who reported similar finding in Turkish population. It is also consistent with the published functional studies that reported some association between XRCC1 Arg399Gln polymorphism and markers of DNA damage. In these studies, it has been shown that the XRCC1 399Gln polymorphic variant is associated with higher levels of DNA adducts, somatic mutations, micronuclei, sister chromatid exchanges and chromosomal damages. In patients with ESRD, DNA damage has been shown by numerous biomarkers, such as the analysis of sister chromatid exchange and chromosomal aberrations,Citation37 comet assay (single-cell gel electrophoresis) in peripheral lymphocytes,Citation21 8-hydroxy 2-deoxyguanosine content in leukocytesCitation38 and mitochondrial DNA deletions in skeletal muscle. Moreover, the XRCC1 399Gln gene variant has been associated with arthrosclerotic coronary artery disease, schizophrenia, pterygium, cataracts and systemic lupus erythematosusCitation3,Citation39–41 as well as various cancer types such as breast, lung, prostate, renal, carcinomas of head and neck, stomach, colon and acute lymphoblastic leukemia.Citation42–49
Combinations of common genetic polymorphisms may increase or decrease the susceptibility to certain diseases.Citation50 To investigate the presence of such an effect, we also made an association analysis between genotype combinations and ESRD. It was found that combination of Arg/Gln or Gln/Gln genotypes of XRCC1 Arg399Gln polymorphism (at least one Gln carriers) with Asp/Asn or Asn/Asn genotypes of XPDAsp312Asn polymorphism or Lys/Gln or Gln/Gln genotypes of XPD Lys751Gln polymorphism may increase the risk of ESRD development. These results are in agreement with Trabulus et al. study.Citation35 We did not find any association between combination of Asp/Asp genotype of XPDAsp312Asn polymorphism with Arg/Gln or Gln/Gln genotypes of XRCC1 Arg399Gln polymorphism and ESRD risk. This is in contrast to the Trabulus et al.Citation35 findings, who reported a positive association between these combined genotypes and ESRD in Turkish population. Furthermore, our results revealed that the combined Lys/Lys genotype of XPD Lys751Gln and Arg/Gln or Gln/Gln genotypes of XRCC1 Arg399Gln did not increase with the risk of ESRD. Lys/Lys genotype of XPD Lys751Gln polymorphism has been reported previously to have a protective effect against certain diseases.Citation51,Citation52
Haplotype association of Gln allele of XRCC1 Arg399Gln with either Asn allele of XPDAsp312Asn or Gln allele of XPD Lys751Gln polymorphism may also increase the risk of ESRD development. Furthermore, haplotypes association of the three codons revealed that association of Gln allele of XRCC1 399 with the Asn allele of XPD 312 and Gln allele of XPD 751 was significantly associated with the development of ESRD.
The limitations of this study are the small population size. The study measured only the genetic variants of XPD and XRCC1. Other risk factors of ESRD were not determined and hence multivariate regression analysis was not done.
In conclusion, our study demonstrated that XRCC1 Arg399Gln polymorphism may contribute to individual susceptibility to ESRD. Thus, BER genes are suggested to be used as a predictive factor for ESRD. However, further studies are needed to evaluate the influence of their polymorphisms on the risk of ESRD.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- Gutiérrez-Amavizca BE, Orozco-Castellanos R, Ortíz-Orozco R, et al. Contribution of GSTM1, GSTT1, and MTHFR polymorphisms to end-stage renal disease of unknown etiology in Mexicans. Indian J Nephrol. 2013;23(6):438–443
- Schupp N, Stopper H, Rutkowski P, et al. Effect of different hemodialysis regimens on genomic damage in end-stage renal failure. Semin Nephrol. 2006;26:28–32
- Guven M, Guven GS, Oz E, et al. DNA repair gene XRCC1 and XPD polymorphisms and their association with coronary artery disease risks and micronucleus frequency. Heart Vessels. 2007;22:355–360
- Maisonneuve P, Agodoa L, Gellert R, et al. Cancer in patients on dialysis for end-stage renal disease: An international collaborative study. Lancet. 1999;354:93–99
- Tepel M, Echelmeyer M, Orie NN, Zidek W. Increased intracellular reactive oxygen species in patients with end-stage renal failure: Effect of hemodialysis. Kidney Int. 2000;58:867–872
- Floccari F, Aloisi C, Crascì E. Oxidative stress and uremia. Med Res Rev. 2005;25:473–486
- Locatelli F, Canaud B, Eckardt KU, Stenvinkel P, Wanner C, Zoccali C. Oxidative stress in end-stage renal disease: An emerging threat to patient outcome. Nephrol Dial Transplant. 2003;18:1272–1280
- Cadet J, Douki T, Ravanat JL. Measurement of oxidatively generated base damage in cellular DNA. Mutat Res. 2011;711:3–12
- Stern MC, Siegmund KD, Conti DV, Corral R, Haile RW. XRCC1, XRCC3, and XPD polymorphisms as modifiers of the effect of smoking and alcohol on colorectal adenoma risk. Cancer Epidemiol Biomarkers Prev. 2006;15:2384–2390
- Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58:604–608
- Lunn RM, Langlois RG, Hsieh LL, Thompson CL, Bell DA. XRCC1 polymorphisms: Effects on aflatoxin B1-DNA adducts and glycophorin A variant frequency. Cancer Res. 1999;59:2557–2561
- Benhamou S, Sarasin A. ERCC2/XPD gene polymorphisms and cancer risk. Mutagenesis 2002;17:463–469
- Catana A, Popp RA, Pop M, Porojan MD, Petrisor FM and Pop IV. Genetic polymorphism of DNA repair gene ERCC2/XPD (Arg 156 Arg) (A22541C) and lung cancer risk in Northern Romania. Rev Romănă Med Lab. 2012;20(2):157–161
- Procopciuc LM, Osian G. Interaction between lifestyle factors and the XRCC1, XPD, and XRCC3 genetic variations modulates the risk for sporadic colorectal cancer. Rev Romănă Med Lab. 2014;22(1):129–141
- Yeh CC, Sung FC, Tang R, Chang-Chieh CR, Hsieh LL. Polymorphisms of the XRCC1, XRCC3, and XPD genes, and colorectal cancer risk: A case-control study in Taiwan. BMC Cancer. 2005;5:12
- Güven M, Unal M, Batar B, et al. Polymorphisms of DNA repair genes XRCC1 and XPD and risk of primary open angle glaucoma (POAG). Mol Vis. 2007;13:12–17
- Unal M, Güven M, Batar B, Ozaydin A, Sarici A, Devranoğlu K. Polymorphisms of DNA repair genes XPD and XRCC1 and risk of cataract development. Exp Eye Res. 2007;85:328–334
- Sorour A, Ayad MW, Kassem H. The genotype distribution of the XRCC1, XRCC3, and XPD DNA repair genes and their role for the development of acute myeloblastic leukemia. Genet Test Mol Biomarkers. 2013;17(3):195–201
- Özcan A, Pehlivan M, Tomatir AG, et al. Polymorphisms of the DNA repair gene XPD (751) and XRCC1 (399) correlates with risk of hematological malignancies in Turkish population. Afr J Biotechnol. 2011;10(44):8860–8870
- Shi JY, Ren ZH, Jiao B, et al. Genetic variations of DNA repair genes and their prognostic significance in patients with acute myeloid leukemia. Int J Cancer. 2011;128(1):233–238
- Caldecott KW. XRCC1 and DNA strand break repair. DNA Repair (Amst). 2003;2:955–969
- El-Din MS, Raslan H, Abdel-Hamid S, Makhlouf M. Detection of XRCC1 gene polymorphisms in Egyptian patients with acute myeloid leukemia. Comp Clin Pathol. 2012;21(5):505–513
- Kuptsova N, Kopecky KJ, Godwin J, et al. Polymorphisms in DNA repair genes and therapeutic outcomes of AML patients from SWOG clinical trials. Blood. 2007;109(9):3936–3944
- Joseph T, Kusumakumary P, Chacko P, Abraham A, Pillai MR. DNA repair gene XRCC1 polymorphisms in childhood acute lymphoblastic leukemia. Cancer Lett. 2005;217(1):17–24
- Meza-Espinoza JP, Peralta-Leal V, Gutierrez-Angulo M, et al. XRCC1 polymorphisms and haplotypes in Mexican patients with acute lymphoblastic leukemia. Genet Mol Res. 2009;8(4):1451–1458
- Ganster C, Neesen J, Zehetmayer S, et al. DNA repair polymorphisms associated with cytogenetic subgroups in B cell chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2009;48:760–767
- Liu J, Song B, Wang Z, et al. DNA repair gene XRCC1 polymorphisms and non-Hodgkin lymphoma risk in a Chinese population. Cancer Genetics Cytogenet. 2009;191(2):67–72
- Smedby KE, Lindgren CM, Hjalgrim H, et al. Variation in DNA repair genes ERCC2, XRCC1, and XRCC3 and risk of follicular lymphoma. Cancer Epidemiol Biomarkers Prev. 2006;15(2):258–265
- Bsnescu C, Trifa AP, Demian S, et al. Polymorphism of XRCC1, XRCC3, and XPD genes and risk of chronic myeloid leukemia. BioMed Res Int. 2014. [Epub ahead of print]
- Ladiges W, Wiley J, MacAuley A. Polymorphisms in the DNA repair gene XRCC1 and age-related disease. Mech Ageing Dev. 2003;124:27–32
- Yuan T, Deng S, Liu H, Liu M, Chen P. Relationship between XRCC1 and XPD polymorphisms and the risk of the development of hepatocellular carcinoma: A case-control study. Exp Ther Med. 2012;4(2):285–290
- Guo LY, Jin XP, Niu W, Li XF, Liu BH, Wang YL. Association of XPD and XRCC1 genetic polymorphisms with hepatocellular carcinoma risk. Asian Pac J Cancer Prev. 2012;13(9):4423–4426
- Batar B, Güven M, Bariş S, Celkan T, Yildiz I. DNA repair gene XPD and XRCC1 polymorphisms and the risk of childhood acute lymphoblastic leukemia. Leuk Res. 2009;33(6):759–763
- Benhamou S, Sarasin A. Variability in nucleotide excision repair and cancer risk: A review. Mutat Res. 2000;462:149–158
- Trabulus S, Guven GS, Altiparmak MR, et al. DNA repair XRCC1 Arg399Gln polymorphism is associated with the risk of development of end-stage renal disease. Mol Biol Rep. 2012;39(6):6995–7001
- Coin F, Marinoni JC, Rodolfo C, Fribourg S, Pedrini AM, Egly JM. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat Genet. 1998;20:184–188
- Nazarkina ZK, Khodyreva SN, Marsin S, Lavrik OI, Radicella JP. XRCC1 interactions with base excision repair DNA intermediates. DNA Repair (Amst). 2007;6:254–264
- Battershill JM, Burnett K, Bull S. Factors affecting the incidence of genotoxicity biomarkers in peripheral blood lymphocytes: Impact on design of biomonitoring studies. Mutagenesis. 2008;23:423–437
- Chiang CC, Tsai YY, Bau DT, et al. Pterygium and genetic polymorphisms of the DNA repair enzymes XRCC1, XPA, and XPD. Mol Vis. 2010;16:698–704
- Padma G, Mamata M, Reddy KR, Padma T. Polymorphisms in two DNA repair genes (XPD and XRCC1)—Association with age related cataracts. Mol Vis. 2011;17:127–133
- Warchoł T, Mostowska A, Lianeri M, Lącki JK, Jagodziński PP. XRCC1 Arg399Gln gene polymorphism and the risk of systemic lupus erythematosus in the polish population. DNA Cell Biol. 2012;31:50–56
- Silva SN, Moita R, Azevedo AP, et al. Menopausal age and XRCC1 gene polymorphisms: Role in breast cancer risk. Cancer Detect Prev. 2007;31:303–309
- Butkiewicz D, Rusin M, Sikora B, Lach A, Chorązy M. An association between DNA repair gene polymorphisms and survival in patients with resected non-small cell lung cancer. Mol Biol Rep. 2011;38:5231–5241
- Berhane N, Sobti RC, Mahdi SA. DNA repair genes polymorphism (XPG and XRCC1) and association of prostate cancer in a north Indian population. Mol Biol Rep. 2012;39(3):2471–2479
- Sturgis EM, Castilli EJ, Li L, et al. Polymorphisms of DNA repair gene XRCC1 in squamous cell carcinoma of the head and the neck. Carcinogenesis. 1999;20:2125–2129
- Duarte MC, Colombo J, Rossit AR, et al. Polymorphisms of DNA repair genes XRCC1 and XRCC3, interaction with environmental exposure and risk of chronic gastritis and gastric cancer. World J Gastroenterol. 2005;11:6593–6600
- Abdel-Rahman S, Soliman AS, Bondy ML, et al. Inheritance of the 194Trp and the 399Gln variant alleles of the DNA repair gene XRCC1 are associated with increased risk of early-onset colorectal carcinoma in Egypt. Cancer Lett. 2000;159:79–86
- Tumer TB, Yilmaz D, Tanrikut C, Sahin G, Ulusoy G, Arinc E. DNA repair XRCC1 Arg399Gln polymorphism alone, and in combination with CYP2E1 polymorphisms significantly contribute to the risk of development of childhood acute lymphoblastic leukemia. Leuk Res. 2010;34:1275–1281
- Hirata H, Hinoda Y, Matsuyama H, et al. Polymorphisms of DNA repair genes are associated with renal cell carcinoma. Biochem Biophys Res Commun. 2006;342:1058–1062
- Görgün E, Güven M, Unal M, et al. Polymorphisms of the DNA repair genes XPD and XRCC1 and the risk of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2010;51:4732–4737
- Gao W, Romkes M, Zhong S, et al. Genetic polymorphisms in the DNA repair genes XPD and XRCC1, p53 gene mutations and bladder cancer risk. Oncol Rep. 2010;24:257–262
- Huang CG, Iv GD, Liu T, Liu Q, Feng JG, Lu XM. Polymorphisms of COMT and XPD and risk of esophageal squamous cell carcinoma in a population of Yili Prefecture, in Xinjiang, China. Biomarkers. 2011;16:37–41