Abstract
Background: The mitochondrial displacement loop (D-loop) is known to accumulate mutations and SNPs at a higher frequency than other regions of mitochondrial DNA (mtDNA). We had identified chronic kidney disease (CKD) risk-associated SNPs in the D-loop of CKD patients previously. In this study, we investigated the association of SNPs in the D-loop of mtDNA with the kidney survival of CKD. Methods: The D-loop region of mtDNA was sequenced for 119 CKD patients from the inpatient of the Fourth Hospital of Hebei Medical University. The Kaplan–Meier method was used to identify disease outcome-associated SNPs in the D-loop of CKD patients. The Cox proportional hazards model was used to identify risk factors for the kidney survival of CKD. Results: In the present study, we identified 20 SNPs with a frequency higher than 5% and assessed the relationship of these SNPs with kidney survival time in CKD patients, a SNP of 146 was identified by log-rank test for statistically significant prediction of the kidney survival time. In an overall multivariate analysis, allele 146 was identified as an independent predictor of kidney survival time in CKD patients. The survival time of kidney in the CKD patients with 146C was significantly shorter than that of kidney in CKD patients with 146T (relative risk, 2.336; 95% CI, 1.319–3.923; p = 0.001). Conclusion: SNPs in the D-loop can predict the kidney survival of CKD patients. Analysis of genetic polymorphisms in the mitochondrial D-loop can help to identify CKD patient subgroup at high risk of a poor disease outcome.
Introduction
Chronic kidney disease (CKD) is becoming a major public health concern. An estimated 23 million people in the United States (11.5% of the adult population) have CKD and are at increased risk for cardiovascular events and progression to kidney failure.Citation1,Citation2 CKD is also highly prevalent in developing countries.Citation3,Citation4 Recently, based on a report published on the Lancet in 2012, China had a higher incidence and prevalence rate of CKD patients, estimating the overall prevalence of CKD to be 10.8%.Citation5 Although there are proven therapies to improve outcomes in patients with CKD, these therapies may also cause harm and add cost. Clinical decision making for CKD is still challenging due to the variability in rates of disease progression and the competing risk of cardiovascular mortality.Citation6,Citation7 The survival period of kidney was equivalent to the time from diagnosed CKD to start dialysis treatment or receive a renal transplant.Citation8 Currently, there are no widely accepted predictive instruments for CKD progression, it is vital to develop new approaches for the kidney survival time prediction in the patients with CKD.
CKD can result from a variety of ecologically distinct causes, including infectious glomerulonephritis, renal vascular, genetic alterations, autoimmune disease and oxidative stress.Citation9 Increased oxidative stress has been observed in CKD patients before the hemodialysis.Citation10 During the pathogenesis of CKD, ROS play important role in regulating the cell signaling pathways of nuclear factor kappa B (NF-кB), which can promote renal cell apoptosis and senescence and decrease regenerative ability of cells in the kidney.Citation11 Mitochondria are major intracellular sources of ROS and highly susceptible to oxidative damage.Citation12,Citation13 Hence, we aimed to investigate the possible relationship between the kidney survival time and mitochondrial DNA (mtDNA) mutations.
The human mitochondrial genome is 16 kb circular double-stranded DNA molecule. It contains 12 coding genes for engaged in respiration and oxidative phosphorylation, two rRNAs, and a set of 22 tRNAs that are essential for the protein synthesis in mitochondria.Citation14 mtDNA is believed to be more susceptible to DNA damage and acquires mutations at a higher rate than nuclear DNA, due to high levels of ROS, a lack of protective histones and a limited capacity for DNA repair in the mitochondria.Citation15,Citation16 mtDNA contains a non-coding region that includes a unique displacement loop (D-loop) that controls replication and transcription of mtDNA, because it contains the initial site of heavy chain replication and the promoters for heavy and light chain transcription. The D-loop is highly polymorphic, and some polymorphisms are associated with aging,Citation17 coronary artery disease,Citation18 and cancers.Citation19,Citation20 However, few of these studies focused on the association of SNPs in the mtDNA D-loop region with the kidney survival time in CKD patients.
The D-Loop contains a length of 1122 bps (nucleotide 16,024–16,569 and 1–576) refer to mitochondria database http://www.mitomap.org. We identified 20 SNPs with a frequency higher than 5% and 6 SNPs (73A\G, 146T\C, 150C\T, 194C\T, 195T\C and 310T\C) associated with the risk of CKD.Citation21 In this study, we assessed the prediction power of these SNPs on the kidney survival time in CKD patients.
Methods
Tissue specimens and DNA extraction
This study was a prospective trial. Blood samples were obtained from 119 CKD patients from the inpatient of the Fourth Hospital of Hebei Medical University between 2002 and 2008. The presence of CKD was defined in accordance with the 2002 National Kidney Foundation Kidney Disease Outcomes Quality Initiative (K-DOQI) classificationCitation22 on the basis of an estimated glomerular filtration rate (eGFR) of less than 60 mL/min/1.73 m2 for at least 90-days. Laboratories in China report eGFR using the four-variable modified diet in renal disease formula.Citation23 The etiologies of CKD in these patients were all chronic glomerulonephritis, including membranous nephropathy (n = 60, 50.4%), non-membranous nephrology (n = 59, 49.6%). The survival period of kidney was equivalent to the time from diagnosed CKD to ESRD. The ESRD was defined as starting dialysis treatment or receiving a renal transplant.Citation8 Total DNA was extracted using a Wizard Genomic DNA extraction kit (Promega, Madison, WI) and stored at −20 °C. Phone calls and follow-up visits to the CKD patients were used to obtain information on patients’ kidney survival status. The study was approved by the Human Tissue Research Committee of the Fourth Hospital of Hebei Medical University. All patients provided written informed consent for the collection of samples and subsequent analysis.
PCR amplification and sequence analysis
The forward primer 5′-CCCCATGCTTACAAGCAAGT-3′ (nucleotide 16,190-16,209) and reverse primer 5′-GCTTTGAGGAGGTAAGCTAC-3′ (nucleotide 602-583) were used for amplification of a 982-bp product from the mtDNA D-loop region. PCR was performed according to the protocol of PCR Master Mix Kit (Promega) and purified prior to sequencing. The PCR condition consisted of one incubation of 2 min at 95 °C, followed by 35 cycles of a 30 s denaturation at 95 °C, a 30 s an annealing at 55 °C, and a 45 s extension at 72 °C, and a final extension at 72 °C for 5 min. Cycle sequencing was performed with the Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystem, Foster City, CA) and the products were then separated on the ABIPRISM Genetic Analyzer 3100 (Applied Biosystem). Polymorphisms were confirmed by repeated analyses from both strands.
Statistical analysis
The kidney survival curve was calculated using the Kaplan–Meier method, and compared with the log-rank test. Multivariate survival analysis was performed using a Cox proportional hazards model. The χ2 test was used to analyze dichotomous values, such as the presence or absence of an individual SNP in CKD patients. All statistical analyses were performed using the SPSS17.0 software (SPSS Company, Chicago, IL). For all the statistical tests, p < 0.05 was considered statistically significant.
Results
A total of 119 CKD patients were enrolled in this study. At baseline, the mean age of 119 CKD patients was 55.6 ± 14.2 years, and 56.3% were male (). The mean eGFR was 81.2 ± 12.4 mL/min/1.73 m2, with 79.8% (n = 95) of them having an eGFR <60 mL/min/1.73 m2. All participants had an eGFR >30 mL/min/1.73 m2. A review was conducted every six months over a five-year period. Seventeen of these patients were lost to follow-up. Those patients lost to follow up during this time period were as follows: three patients in Year 1; four patients in Year 2; four patients in Year 3; five patients in Year 4; one patient in Year 5. The remaining 102 patients were assessed. The relationship between the data collected during the 5-year follow-up and patients’ clinical characteristics were analyzed using the Kaplan–Meier method and the log-rank test. Age, body mass index (BMI) and mean article pressure (MAP) were not statistically significant predictors of the kidney survival time in CKD patients. Sex, smoking classification and eGFR of the starting point were correlated with the kidney survival time of the progressive kidney disease (). The survival rates of kidney in female CKD patients were higher than those of kidney in male CKD patients. Smoking habit showed an association with survival rates of kidney in CKD patients when the survival time of kidney in smoking CKD patients was lower than those of kidney in non-smoking CKD patients. The survival rates of kidney in CKD patients with eGFR <60 mL/min/1.73 m2 was lower than those of kidney in CKD patients with eGFR ≥60 mL/min/1.73 m2. These data demonstrate that gender, smoking habit and the eGFR of baseline were the risk factors for the survival time of kidney in the CKD patients.
Table 1. Baseline characteristics of the CKD participants.
Table 2. Univariate analysis of clinical characteristics associated with the kidney survival time in chronic kidney disease patients.
The correlation between the mtDNA genotype and the kidney survival was compared. SNPs with a minor allele frequency of <5% were excluded; thus, a remaining 20 SNPs were used for further analysis. CKD patients were divided into two groups on the basis of their genotype at each SNP site, the kidney survival curve was plotted using the Kaplan–Meier method for all CKD patients at these sites. A dramatic difference in kidney survival rate was found for allele 146 (). The rare allele 146C was associated with a significantly shorter (p = 0.001) length of kidney survival (). The allele 146 also belong to CKD risk associated SNPs we identified before. We used the multivariate analysis by the Cox proportional hazards model with the prediction factors, including one SNP, gender, smoking and eGFR. As shown in , the 146 alleles were identified as independent predictors for the kidney survival time of progressive kidney disease. The survival time of kidney in the CKD patients with the rare allele 146C genotype was significantly shorter than that of kidney in CKD patients with the frequent allele 146T (relative risk, 2.336; 95% CI, 1.319–3.923; p = 0.001) at the 146 site. The eGFR was also an independent predictor for the kidney survival time of progressive kidney disease. The gender and smoking were excluded in the multivariate analysis by the Cox proportional hazards model. We also further analyzed the relationship of ESRD risk factors between 146C and 146T case, the result showed that there was no difference between 146C and 146T in age, gender, smoking, BMI, MAP, etiologies of CKD and eGFR (). These data demonstrated the strong prediction power of nucleotide 146 on the kidney survival time of progressive kidney disease.
Figure 1. Survival curve of kidney according to the nucleotide at position 146 in D-loop of the chronic kidney disease patients.
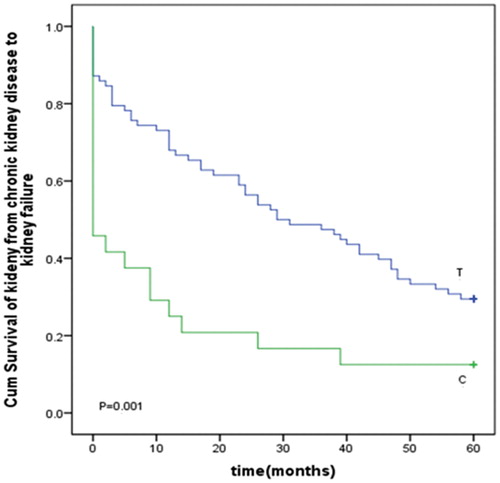
Table 3. Multivariate analysis of prognostic factors associated with the kidney survival time in chronic kidney disease patients by the Cox proportional hazards model.
Table 4. The relationship of clinical characteristics with the frequency of allele 146.
Discussion
Selected SNPs in the mtDNA D-loop region have been examined for the ability to predict disease risk in CKD.Citation21 In our previous case-control study published on Renal Failure,Citation21 we found that the SNPs in the mitochondrial DNA D-loop region were associated with the risk of CKD. The present study has extended those analyses to determine the kidney survival-associated SNPs in a continuous sequence of mtDNA between nucleotides 16,190 and 583. One SNP, 146T/C, was identified by the log-rank test for the association with the kidney survival. Multivariate survival analysis identified 146T/C as an independent prediction marker for the kidney survival time. We identified six SNPs as the CKD risk SNPs including 73G, 146C, 150T, 194T, 195C and 310C.Citation21 It is interesting that the allele 146 also belong to these CKD risk SNPs. Nucleotide 146 is located in hypervariable segment 2 (HV2). The functional significance of HV segments, mutational hotspots at which germ line and mtDNA mutations preferentially occur,Citation24 is not known, but our data suggest that these SNPs are potential genetic markers for the kidney survival time. Minor alleles of 146C are associated with a dramatically shorter period of kidney survival and the kidney survival curve decreased rapidly in CKD patients with these alleles (). The 146 site is hypervariable; Alzheimer’s disease and Parkinson’s disease have been identified as having mutations at this site.Citation25,Citation26 Further study is needed to solve the mystery of this allele.
The D-loop region of mtDNA is important for the regulation of mitochondrial genome replication and expression. SNPs in the D-loop region might affect mtDNA replication and lead to electron transport chain alteration, which is responsible for the release of highly ROS and could contribute to the nuclear genome damage.Citation27,Citation28 The ROS may promote kidney disease by activating the signaling pathways of NF-кB.Citation11 NF-кB is activated by ROS and upregulates of ROS-induced granulocyte macrophage-colony-stimulating factor (GM-CSF), which can enhance glomerular inflammation and induce mesangial proliferation.Citation11
Consistent with earlier reports, our data demonstrated smoking and gender were associated with the CKD progression,Citation29–33 which confirmed the validity of our data. The mechanism explained the relationship between smoking and CKD progression is that smoking contribute to oxidative stress, which plays important role in the development and progression of CKD.Citation34 The relationship between gender and CKD progression suggests that sex hormones may be important determinants of the greater susceptibility of males to progressive kidney injury.
Conclusion
In conclusion, SNPs in the mtDNA D-loop were found to be independent prognostic markers for the kidney survival time in CKD patients. The analysis of genetic polymorphisms in the D-loop might help to identify CKD patient subgroups at high risk for a disease outcome, thereby helping to refine therapeutic decisions in CKD patients.
Declaration of interest
All of the authors disclose any financial, consulting and personal relationships with other people or organizations that could influence the author’s work.
This work was supported by the project of the Hebei Natural Science Fund (H2012206157) and the project of the Hebei Major Medical Science (GL2011-51).
References
- Coresh J, Selvin E, Stevens LA, et al. Prevalence of chronic kidney disease in the United States. JAMA. 2007;298:2038–2047
- Go AS, Chertow GM, Fan D, McCulloch CE, Hsu CY. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N Engl J Med. 2004;351:1296–1305
- Nugent RA, Fathima SF, Feigl AB, Chyung D. The burden of chronic kidney disease on developing nations: A 21st century challenge in global health. Nephron Clin Pract. 2011;118:c269–c277
- Eknoyan G, Lameire N, Barsoum R, et al. The burden of kidney disease: Improving global outcomes. Kidney Int. 2004;66:1310–1314
- Yano Y, Fujimoto S, Asahi K, Watanabe T. Prevalence of chronic kidney disease in China. Lancet. 2012;380:213–214; author reply 214–216
- Keane WF, Zhang Z, Lyle PA, et al. Risk scores for predicting outcomes in patients with type 2 diabetes and nephropathy: The RENAAL study. Clin J Am Soc Nephrol. 2006;1:761–767
- Keith DS, Nichols GA, Gullion CM, Brown JB, Smith DH. Longitudinal follow-up and outcomes among a population with chronic kidney disease in a large managed care organization. Arch Intern Med. 2004;164:659–663
- Schrier RW, McFann KK, Johnson AM. Epidemiological study of kidney survival in autosomal dominant polycystic kidney disease. Kidney Int. 2003;63:678–685
- Lopez-Novoa JM, Martinez-Salgado C, Rodriguez-Pena AB, Lopez-Hernandez FJ. Common pathophysiological mechanisms of chronic kidney disease: Therapeutic perspectives. Pharmacol Ther. 2010;128:61–81
- Himmelfarb J, Stenvinkel P, Ikizler TA, Hakim RM. The elephant in uremia: Oxidant stress as a unifying concept of cardiovascular disease in uremia. Kidney Int. 2002;62:1524–1538
- Greiber S, Muller B, Daemisch P, Pavenstadt H. Reactive oxygen species alter gene expression in podocytes: Induction of granulocyte macrophage-colony-stimulating factor. J Am Soc Nephrol. 2002;13:86–95
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13
- Nishikawa T, Edelstein D, Du XL, et al. Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature. 2000;404:787–790
- Lee HC, Yin PH, Lin JC, et al. Mitochondrial genome instability and mtDNA depletion in human cancers. Ann N Y Acad Sci. 2005;1042:109–122
- DiMauro S, Schon EA. Mitochondrial DNA mutations in human disease. Am J Med Genet. 2001;106:18–26
- Beal MF. Mitochondria, free radicals, and neurodegeneration. Curr Opin Neurobiol. 1996;6:661–666
- Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science. 1999;286:774–779
- Mueller EE, Eder W, Ebner S, et al. The mitochondrial T16189C polymorphism is associated with coronary artery disease in Middle European populations. PLoS One. 2011;6:e16455
- Zhang R, Wang R, Zhang F, et al. Single nucleotide polymorphisms in the mitochondrial displacement loop and outcome of esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2010;29:155
- Guo Z, Yang H, Zhang F, Zhang R, Wang C. Single nucleotide polymorphisms in the mitochondrial displacement loop and age-at-onset of esophageal squamous cell carcinoma. Oncol Lett. 2012;3:482–484
- Bai Y, Guo Z, Xu J, et al. Association of sequence polymorphism in the mitochondrial D-loop with chronic kidney disease. Ren Fail. 2014;36(5):781–784
- Levey AS, Coresh J, Balk E, et al. National Kidney Foundation practice guidelines for chronic kidney disease: Evaluation, classification, and stratification. Ann Intern Med. 2003;139:137–147
- Levey AS, Bosch JP, Lewis JB, Greene T, Rogers N, Roth D. A more accurate method to estimate glomerular filtration rate from serum creatinine: A new prediction equation. Modification of Diet in Renal Disease Study Group. Ann Intern Med. 1999;130:461–470
- Stoneking M. Hypervariable sites in the mtDNA control region are mutational hotspots. Am J Hum Genet. 2000;67:1029–1032
- Takasaki S. Mitochondrial SNPs associated with Japanese centenarians, Alzheimer's patients, and Parkinson's patients. Comput Biol Chem. 2008;32:332–337
- Vives-Bauza C, Andreu AL, Manfredi G, et al. Sequence analysis of the entire mitochondrial genome in Parkinson's disease. Biochem Biophys Res Commun. 2002;290:1593–1601
- Bandy B, Davison AJ. Mitochondrial mutations may increase oxidative stress: Implications for carcinogenesis and aging? Free Radic Biol Med. 1990;8:523–539
- Hervouet E, Simonnet H, Godinot C. Mitochondria and reactive oxygen species in renal cancer. Biochimie. 2007;89:1080–1088
- Nagasawa Y, Yamamoto R, Rakugi H, Isaka Y. Cigarette smoking and chronic kidney diseases. Hypertens Res. 2012;35:261–265
- Daumit GL, Hermann JA, Powe NR. Relation of gender and health insurance to cardiovascular procedure use in persons with progression of chronic renal disease. Med Care. 2000;38:354–365
- Hsu CY, Iribarren C, McCulloch CE, Darbinian J, Go AS. Risk factors for end-stage renal disease: 25-year follow-up. Arch Intern Med. 2009;69:342–350
- Neugarten J, Golestaneh L. Gender and the prevalence and progression of renal disease. Adv Chronic Kidney Dis. 2013;20:390–395
- Chen JB, Yang YH, Lee WC, et al. Sequence-based polymorphisms in the mitochondrial D-loop and potential SNP predictors for chronic dialysis. PLoS One. 2012;7:e41125
- Zalba G, Fortuno A, Diez J. Oxidative stress and atherosclerosis in early chronic kidney disease. Nephrol Dial Transplant. 2006;21:2686–2690

