Abstract
Aim: The investigations into the association between the receptor for advanced glycation end products (RAGE) gene -374T/A, -429T/C polymorphisms and diabetic nephropathy (DN) in several case–control studies have rendered conflicting results. To shed light on these inconclusive findings, a meta-analysis of all the eligible studies relating these two polymorphisms to the risk of DN was conducted. Methods: The databases were searched for relevant articles up to July 2014. A pooled estimate of the genetic association, the heterogeneity between studies, and the publication bias were investigated. Results: Eight studies with 1725 cases and 1857 controls were enrolled in -374T/A polymorphism analysis. The main analysis indicated no association for the allele contrast, the recessive model and the dominant model. Subgroup analyses in Caucasians and in type 2 diabetes also showed no association between -374T/A polymorphism and DN. Five studies with 1019 cases and 792 controls were enrolled in -429T/C polymorphism analysis. The main analysis revealed heterogeneity and no association for the allele contrast and the dominant model. However, the recessive model for -429C allele diminished the heterogeneity and showed a marginal association overall [fixed-effects OR = 2.83 (1.33–6.00) and random effects OR = 2.50 (1.00–6.24), respectively]. Conclusions: Our meta-analysis indicated that the RAGE gene -429CC genotype might be a risk factor for DN in patients with type 2 diabetes.
Introduction
Diabetic nephropathy (DN) is a frequent chronic micro-vascular complication of both type 1 and type 2 diabetes mellitus (DM), and is the primary cause of end-stage renal failure.Citation1 Although severity and duration of diabetes are strong determinants of DN, the etiology of DN is multi-factorial and involves both environmental and genetic factors.Citation2,Citation3 Family clustering, heterogeneity in the onset and progression and results of segregation studies indicated that a genetic predisposition is implicated in the pathogenesis of DN.Citation4
The receptor for advanced glycation end products (RAGE) is a multi-ligand member of the immunoglobulin superfamily of cell-surface molecules,Citation5 that was first described as receptor for advanced glycation end products (AGEs) occurring in diabetes and at sites of oxidant stress in tissues.Citation6,Citation7 Sustained interaction of AGEs-RAGE may trigger RAGE-dependent cellular activation, induce oxidative stress, and promote inflammatory-proliferative responses leading to vascular dysfunction.Citation8–10 The up-regulation and pathogenic effects of RAGE in vascular disease make the RAGE gene an attractive candidate gene for diabetic vascular complication.
RAGE gene is located on chromosome 6p21.3 in MHC Class III region and it is composed of 11 exons, a 3′-UTR (untranslated region) and a 5′ flanking region.Citation11 Genetic studies have identified that approximate 30 polymorphisms occur in the RAGE gene.Citation9 Of all of the RAGE gene polymorphisms identified, two functional polymorphisms, namely -374T/A (rs1800624) and -429T/C (rs1800625) variants in the RAGE gene promoter region, have been shown to increase transcription activity in vitro,Citation12 and therefore have attracted considerable interest. However, case–control studies that investigated the association between these two polymorphisms and DN have rendered conflicting results.Citation13–21 To shed light on these inconclusive results, a meta-analysis of all eligible studies relating the RAGE gene -374T/A and -429T/C polymorphisms to the risk of developing DN was conducted.
Materials and methods
Identification and eligibility of relevant studies
All studies published before July 2014 was identified by extended computer-based searches of the electronic databases (PubMed, EMBASE, ISI web of science and Chinese National Knowledge Infrastructure). The search strategy was based on the combination of (“RAGE” or “AGER” or “advanced glycosylation end-product receptor”) and “polymorphism” and (“diabetic nephropathy” or “diabetes complications”). References of retrieved articles were also screened. Abstracts, case reports, editorials and review articles were excluded. Studies included in the meta-analysis had to meet all the following criteria: (a) use an unrelated case–control design, (b) have available genotype or allele frequency, and (c) cases were DN patients and controls were type 1 or type 2 diabetes patients without nephropathy. Studies based on pedigree data were excluded since they investigated linkage and not association.Citation22
Data extraction
Data were extracted by two investigators. The extracted data included information about first author, year of publication, country of origin, ethnicity of study population, clinical characteristics, the genotyping method, the number of cases and controls, the distribution of genotypes and the allele frequencies for both cases and controls. If encountered the conflicting evaluations, and agreement was reached following a discussion; if could not reached agreement, then a third author was consulted to resolve the debate. For articles including subjects of different racial descent or different type of DM, data were extracted separately for each race or each type of DM. In the case of multiple publications of analyses of the same data or overlapping data sets, the publication that reported data from the largest or most recent study was included.
Meta-analysis
The meta-analysis examined the overall association of the allele contrast, the recessive model and the dominant model. All associations were indicated as odds ratios (OR) with the corresponding 95% confidence interval (CI). Based on the individual OR, a pooled OR was estimated. Heterogeneity between studies was tested using the χ2-based Q-statistic. Owing to the low power of the statistic, heterogeneity was considered significant for PQ-value < 0.10.Citation23 The pooled OR was estimated using fixed-effects (Mantel–Haenszel) and random-effects (DerSimonian and Laird) models. Random effects model assumes a genuine diversity in the results of various studies and incorporates a between-study variance into the calculations.Citation23 Therefore, when there is heterogeneity between studies (PQ < 0.10), the pooled OR is estimated using the random effects model.Citation23 Otherwise, the pooled OR is estimated using the fixed-effects model. Statistical power of meta-analysis was calculated with power calculator software PASS (Power and Sample Size). Publication bias was assessed by Egger’s linear regression test.Citation24 Given that Egger’s linear regression test is underpowered, it was considered significant for PE-value < 0.10. In addition to the main analysis, subgroup analyses for the studies of Caucasian descent and type 2 diabetes (studies of Asian and African descent and type 1 diabetes were not included in subgroup analyses because the number of studies did not meet the minimal requirement of four studies) were also performed. The distribution of genotypes in the control group was tested for Hardy–Weinberg equilibrium (HWE) using an exact test.Citation25 Lack of HWE indicates possible genotyping errors and/or population stratification.Citation3 Studies with controls not in HWE were subjected to a sensitivity analysis. Meta-analysis was performed using the Review Manager version 5.3 (Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration) and STATA package version 11.0 (Stata Corporation, College Station, TX). All p-values were two-sided.
Results
Eligible studies
The literature review identified 61 titles in the electronic databases that met the search criterion. After review, seven articles investigating the association between the RAGE gene -374T/A polymorphism and DN,Citation13,Citation14,Citation16–19,Citation21 and four articles investigating the association between the -429T/C polymorphism and DN met the inclusion criteria.Citation15,Citation17,Citation18,Citation20 Among them one article about -374T/A polymorphismCitation21 was excluded because it partially overlapped with another studyCitation19 that provided a larger data. In the rest of articles, dos Santos et al.Citation18 described on two ethnic groups (Caucasian- and African-Brazilians) and Lindholm et al.Citation19 provided data about type 1 and type 2 diabetes separately. According to methodology, each race or each type of diabetes in these two articles was treated as a separate study in the meta-analysis. Finally, data from six articles (eight case–control studies) involved -374T/A polymorphism and four articles (five case–control studies) involved -429T/C polymorphism were included in the meta-analysis. A list of details abstracted from the studies included in the meta-analysis is provided in .
Table 1. Characteristics of the case–control studies considered in the meta-analysis.
Summary statistics
In total, the studies for -374T/A polymorphism included 1725 cases and 1857 controls, and the studies for -429T/C polymorphism included 1019 cases and 792 controls. In all studies, two case–control studies provided data only for allele frequencies without genotypes information.Citation18 One study did not describe all genotypes separately; it provided data only for -374T-carriers.Citation14 In all studies, the genotype distribution of control groups was in HWE (p ≥ 0.05). The prevalence of -374A and -429C allele was 27.8% and 13.0% in controls, respectively. The -374A allele had a lower representation in controls of Asians (13.1%) than in controls of Caucasians (29.2%, range: 27.0–37.5%) and Africans (28.0%). The -429C allele was equally represented in controls of Asians (13.2%, range: 12.0–15.4%) as in controls of Caucasians (13.0%, range: 11.0–15.1%) and Africans (12.1%) ().
Table 2. Distribution of the RAGE gene -374T/A and -429T/C genotypes for cases and controls and the allele frequencies.
Main results and subgroup analyses
The main analysis for investigating the association between -374T/A allele A and the risk of developing DN relative to the allele T revealed significant heterogeneity (PQ = 0.03) between the seven studies, and the random effects pooled OR was not significant [RE OR = 1.03 (0.85–1.25)]. In subgroup analysis, the fixed- and random-effects ORs were not significant in Caucasian population and in type 2 diabetes. The recessive model (AA vs. TT+TA) and the dominant model (AA+TA vs. TT) for -374A allele also showed lack of association in main analysis and subgroup analysis (, ).
Figure 1. Random effects odds ratio estimates with the corresponding 95% confidence interval for the allele contrast A versus T of RAGE gene -374T/A polymorphism.
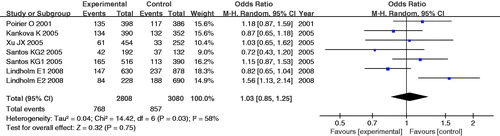
Table 3. Summary of the odds ratio (OR) for genetic contrasts of the association of -374T/A and -429T/C polymorphisms in DN.
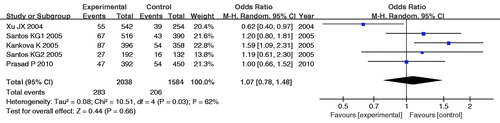
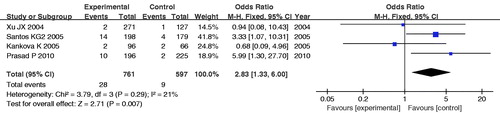
The main analysis for investigating the association of the -429C allele and the risk of DN relative to the -429T allele showed significant heterogeneity (PQ = 0.03) between the five studies, and the pooled OR by random effects was not significant [RE OR = 1.07 (0.78–1.48)]. The dominant model for -429C allele (CC+TC vs. TT) yielded similar pattern of results with the allele contrast. However, the recessive model (CC vs. TT+TC) derived a marginal significant association overall [PQ = 0.29, FE OR = 2.83 (1.33–6.00) and RE OR = 2.50 (1.00–6.24), respectively] (, and ). The statistical power (α = 0.05) for the recessive model of -429T/C polymorphism was 72%.
Publication bias
Egger’s linear regression test indicated that there is no significant publication bias with all available studies both in the comparison of -374 A versus T allele [t = 0.02, PE = 0.98 (−7.22 to 7.36)] and -429 C versus T allele [t = −0.39, PE = 0.72 (−16.86 to 13.20)].
Discussion
Investigation with the RAGE gene -374T/A variant had revealed that the introduction of a T-to-A nucleotide substitution apparently prevents the binding of a nuclear binding factor and the presence of the -374A allele increases the promoter transcription activity in vitro.Citation12 Our meta-analysis for the -374T/A polymorphism included data from six articles (eight case–control studies) and comprised a total of 1725 cases and 1857 controls. No publication bias was detected for all available studies. The main analysis indicated significant between-study heterogeneity and no association of the -374A allele with the risk of DN relative to the -374T allele. Subgroup analyses in Caucasians and in type 2 diabetes also showed no associations for the allele contrast, with similar results in the recessive model and the dominant model. Taken together, these data suggested that there might be no association of RAGE gene -374T/A polymorphism with DN risk. The lack of association in the -374T/A polymorphism analyses could be due to lack of power to detect existing significant association and to other loci that are probably in linkage disequilibrium and that may affect RAGE susceptibility to DN.
RAGE gene -429T/C variant is proximal to the -374T/A variant. Our meta-analysis for the -429T/C polymorphism involved four articles (five case–control studies), which provided 1019 cases and 792 controls. Both cases and controls were patients with type 2 diabetes. No publication bias was detected for all available studies. The overall data showed heterogeneity and no significant association for the allele contrast. The dominant model produced the same pattern of results with the allele contrast. However, the recessive model for -429C allele diminished the heterogeneity and showed a significant association by fixed and random effects. Therefore, these data suggest that the RAGE gene -429CC genotype might contribute to the susceptibility of DN in patients with type 2 diabetes.
It might be argued that our positive results of the meta-analysis could be due to a statistical error caused by the relatively limited available studies, but it does not seem to be the case because in vitro study has observed that the RAGE gene -429T/C polymorphism increase transcriptional levels.Citation12 Moreover, a recent meta-analysis found that the dominant model of -429T/C polymorphism increased the odds of developing coronary artery disease in diabetic patients by 1.22-fold compared with that of non-diabetic patients of 1.07-folds.Citation26 Therefore, this variant also might be involved in the pathogenesis of DN. In addition, the strength of meta-analysis was based on the accumulation of published data giving greater information to detect significant differences. Our meta-analysis has produced results which suggest more research could be warranted.
Some limitations of our meta-analysis should be addressed. First, our subgroup analyses provided data only for type 2 diabetes and Caucasian population, but no sufficient studies available to do stratification analyses in type 1 diabetes, Asian and African population. Future studies should address more in type 1 diabetes and other ethnic groups. Second, although cases and controls of each study were well defined with similar inclusion criteria, there may be factors that were not taken into account that may have at least in part influenced our results.
In conclusion, the present meta-analysis supports an association between the RAGE gene -429T/C polymorphism and DN. The -429CC genotype might be a risk factor for DN in patients with type 2 diabetes. DN is a complex disease with a multi-factorial etiology and thus studies that investigate gene–gene, gene–environment interactions should help further elucidate the genetics of DN.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the article.
References
- Gross JL, de Azevedo MJ, Silveiro SP, Canani LH, Caramori ML, Zelmanovitz T. Diabetic nephropathy: Diagnosis, prevention, and treatment. Diabetes Care. 2005;28:164–176
- Chowdhury TA, Dyer PH, Kumar S, Barnett AH, Bain SC. Genetic determinants of diabetic nephropathy. Clin Sci (Lond). 1999;96:221–230
- Zintzaras E, Uhlig K, Koukoulis GN, Papathanasiou AA, Stefanidis I. Methylenetetrahydrofolate reductase gene polymorphism as a risk factor for diabetic nephropathy: A meta-analysis. J Hum Genet. 2007;52:881–890
- Badal SS, Danesh FR. New insights into molecular mechanisms of diabetic kidney disease. Am J Kidney Dis. 2014;63:S63–S83
- Neeper M, Schmidt AM, Brett J, et al. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J Biol Chem. 1992;267:14998–15004
- Schmidt AM, Vianna M, Gerlach M, et al. Isolation and characterization of two binding proteins for advanced glycosylation end products from bovine lung which are present on the endothelial cell surface. J Biol Chem. 1992;267:14987–14997
- Schmidt AM, Hori O, Brett J, Yan SD, Wautier JL, Stern D. Cellular receptors for advanced glycation end products. Implications for induction of oxidant stress and cellular dysfunction in the pathogenesis of vascular lesions. Arterioscler Thromb. 1994;14:1521–1528
- Chavakis T, Bierhaus A, Al-Fakhri N, et al. The pattern recognition receptor (RAGE) is a counterreceptor for leukocyte integrins: A novel pathway for inflammatory cell recruitment. J Exp Med. 2003;198:1507–1515
- Kalea AZ, Schmidt AM, Hudson BI. RAGE: A novel biological and genetic marker for vascular disease. Clin Sci (Lond). 2009;116:621–637
- Singh VP, Bali A, Singh N, Jaggi AS. Advanced glycation end products and diabetic complications. Korean J Physiol Pharmacol. 2014;18:1–14
- Sugaya K, Fukagawa T, Matsumoto K, et al. Three genes in the human MHC class III region near the junction with the class II: Gene for receptor of advanced glycosylation end products, PBX2 homeobox gene and a notch homolog, human counterpart of mouse mammary tumor gene int-3. Genomics. 1994;23:408–419
- Hudson BI, Stickland MH, Futers TS, Grant PJ. Effects of novel polymorphisms in the RAGE gene on transcriptional regulation and their association with diabetic retinopathy. Diabetes. 2001;50:1505–1511
- Poirier O, Nicaud V, Vionnet N, et al. Polymorphism screening of four genes encoding advanced glycation end-product putative receptors. Association study with nephropathy in type 1 diabetic patients. Diabetes. 2001;50:1214–1218
- Pettersson-Fernholm K, Forsblom C, Hudson BI, Perola M, Grant PJ, Groop PH. The functional -374 T/A RAGE gene polymorphism is associated with proteinuria and cardiovascular disease in type 1 diabetic patients. Diabetes. 2003;52:891–894
- Xu JX, Xu B, Yang MG, et al. -429T/C polymorphism of RAGE gene promoter is associated with type 2 diabetic nephropathy. China J Endocrinol Metab. 2004;20:534–535
- Xu JX, Yang MG, Liu JY, Chang YL, Zhang PW, Chen HX. Association between RAGE gene promoter -374T/A polymorphism and type 2 diabetes and nephropathy. Chin J Prev Contr Chron Non-Commun Dis. 2005;13:39–40
- Kankova K, Stejskalova A, Hertlova M, Znojil V. Haplotype analysis of the RAGE gene: Identification of a haplotype marker for diabetic nephropathy in type 2 diabetes mellitus. Nephrol Dial Transplant. 2005;20:1093–1102
- dos Santos KG, Canani LH, Gross JL, Tschiedel B, Pires Souto KE, Roisenberg I. The -374A allele of the receptor for advanced glycation end products gene is associated with a decreased risk of ischemic heart disease in African-Brazilians with type 2 diabetes. Mol Genet Metab. 2005;85:149–156
- Lindholm E, Bakhtadze E, Cilio C, Agardh E, Groop L, Agardh CD. Association between LTA, TNF and AGER polymorphisms and late diabetic complications. PLoS One. 2008;3:e2546
- Prasad P, Tiwari AK, Kumar KM, et al. Association analysis of ADPRT1, AKR1B1, RAGE, GFPT2 and PAI-1 gene polymorphisms with chronic renal insufficiency among Asian Indians with type-2 diabetes. BMC Med Genet. 2010;11:52
- Lindholm E, Bakhtadze E, Sjogren M, et al. The -374 T/A polymorphism in the gene encoding RAGE is associated with diabetic nephropathy and retinopathy in type 1 diabetic patients. Diabetologia. 2006;49:2745–2755
- Zintzaras E, Ioannidis J. Heterogeneity testing in metanalysis of genome searches. Genet Epidemiol. 2005;28:123–137
- Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634
- Weir B. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sunderland, MA: Sinauer Associates; 1996
- Feng P, Dan H, Nan J, et al. Association of four genetic polymorphisms of AGER and its circulating forms with coronary artery disease: A meta-analysis. PLoS One. 2013;8:e70834