Abstract
ATP-binding cassette transporters are multi-subunit membrane pumps that transport substrates across membranes. While significant in the transport process, transporter architecture exhibits a range of diversity that we are only beginning to recognize. This divergence may provide insight into the mechanisms of substrate transport and homeostasis. Until recently, ABC importers have been classified into two types, but with the emergence of energy-coupling factor (ECF) transporters there are potentially three types of ABC importers. In this review, we summarize an expansive body of research on the three types of importers with an emphasis on the basics that underlie ABC importers, such as structure, subunit composition and mechanism.
Introduction
ABC transporters, present in all organisms, represent one of the largest transmembrane spanning super-families. ABC stands for ATP-binding cassette due to the family's characteristic ability to bind and hydrolyze ATP in order to transport substrates across the lipid bilayer (Higgins, Citation1992). Found in both prokaryotes and eukaryotes, the well-studied ABC exporters, play a fundamental role in biological processes (Dean et al., Citation2001), are associated with multidrug resistance (Eckford & Sharom, Citation2009; Kerr et al., Citation2010; Seeger & van Veen, Citation2009), several human diseases (Dean et al., Citation2001; Fletcher et al., Citation2010) and have been extensively reviewed elsewhere (Al-Shawi, Citation2011). ABC importers play a major role in nutrient uptake, pathogenicity and virulence, making a focus on this diverse set of transporters ever more prevalent. ABC importers found in plants, prokaryotes and archaea are responsible for the transport of vital nutrients across lipid bilayers. In addition, many prokaryotic pathogens utilize ABC importers to circumvent the hosts innate immunes response, which makes this group of transporters of particular interest.
All ABC transporters have a common basic architecture with two nucleotide-binding domains (NBDs) and two transmembrane domains (TMDs). These domains assemble to form the basic unit of an ABC transporter. For ABC exporters, the TMD and NBD domains can be fused in multiple ways resulting in one-, two- or four-chain transporters, where the order of the domains may vary. For importers, the core TMD and NDB subunits are individual chains, which can assemble to form homo- or hetero-dimeric TMDs bound to typically homodimeric NBDs. The NBD is classified by a series of motifs, including the Walker A (P-loop), Walker B and ABC signature motif, which are common in, but not limited to ABC transporters (Higgins et al., Citation1986; Walker et al., Citation1982). These motifs, in conjunction with additional conserved NBD regions (H-loop/switch region, Q-loop), play a role in ATP binding (Schneider & Hunke, Citation1998). The binding and hydrolysis of ATP in the NBDs are thought to control the opening and closing of the TMDs.
Unlike the NBD, the TMD sequence is considerably less conserved. For ABC importers, even the number of helices per TMD subunit can vary from 5 to 10, yielding a total of 10–20 transmembrane helices per transporter (Davidson et al., Citation2008). TMDs of the ABC uptake systems contain the so-called EAA motif (L-loop), a cytoplasmic loop containing two short helices that couple conformational shifts of the NBDs to the TMDs (Saurin & Dassa, Citation1994, Saurin et al., Citation1994). Together, the TMDs and NBDs form a complex to make up the core unit of an ABC transporter. For most ABC importers, substrate is delivered to a core importer complex through a substrate-binding protein (SBP). SBPs bind to a vast array of substrates with affinities in the nanomolar to micromolar range. These proteins are located in the periplasm of gram-negative organisms or tethered to the membrane in gram-positive bacteria. The most recent discovery of ECF or energy coupling factor transporters has revealed a divergence in ABC importers. ECF transporters contain both NBDs and TMDs but do not have an additional SBP for substrate delivery. Instead, a membrane-embedded component of the ECF transporter binds substrate. The overall topology and mechanisms each type of importer uses to transport substrates emphasizes the diversity amongst ABC importers.
Intact prokaryotic structures reveal diversity amongst ABC importers
Structures of ABC importers have revealed a divergence in overall architecture that is not quite understood. To date, 12 high-resolution crystal structures of importers have been solved, all of prokaryotic origin. Based on overall topology and mechanism of transport, several structurally characterized ABC importers fall within the Type I category including: multiple conformations of the maltose transporter MalFGK2 (Chen et al., Citation2013; Khare et al., Citation2009; Oldham & Chen, Citation2011; Oldham et al., Citation2007), methionine transporter MetNI (Kadaba et al., Citation2008) and molybdate transporters ModBC from Archaeoglobus fulgidus (Hollenstein et al., Citation2007) and Methanosarcina acetivorans (Gerber et al., Citation2008). These differ from Type II importers which have been classified based on the structures of the vitamin B12 transporter BtuCD (Locher et al., Citation2002), the heme transporter HmuUV (Woo et al., Citation2012), and the molybdate transporter MolBC (HI1470/1) from Haemophilus influenzae (Pinkett et al., Citation2007). The structural diversity of ECF transporters or Type III ABC importers was revealed with the full length structures of the folate (Xu et al., Citation2013) and the hydroxymethyl pyrimidine (Wang et al., Citation2013) transporters from Lactobacillus brevis.
Previous reviews have covered the conserved aspects of ABC transporters (Davidson et al., Citation2008) or focused on structural characteristics (Biemans-Oldehinkel et al., Citation2006) and the initial division of Type I and II importers (Locher, Citation2009; Oldham et al., Citation2008). Since then much has been added to the field that expands our current understanding of transporter diversity, including ECF transporters, which have been the focus of several recent reviews (Erkens et al., Citation2012; Slotboom, Citation2014; Zhang, Citation2013). This review is a culmination of the recent advances for all three types of ABC importers, with a focus on structures and mechanisms employed to transport substrates across the lipid bilayer.
Type I ABC importers
Structural and biochemical evidence has allowed for the division of the different types of ABC importer. In general, the Type I importers contain fewer transmembrane helices in comparison with Type II. In the absence of SBPs, Type I transporters show negligible levels of ATPase activity; however, activity is stimulated by ligand-loaded SBP (Bao & Duong, Citation2012; Davidson et al., Citation1992). The structurally characterized Type I importers shed light on the diversity of transmembrane domain topology (). The Type I importer MetNI has a minimal set of 5 TM helices from each TMD, which forms a homodimer totaling 10 helices (Kadaba et al., Citation2008). The structures of two molybdate transporters (ModBC homologs) have an additional TM helix per monomer, totaling 12 helices (Gerber et al., Citation2008; Hollenstein et al., Citation2007). Unlike other ABC importers, which form homodimers, the TMDs of maltose transporter form a heterodimer of MalF and MalG, which have 8 and 6 helices, respectively. Despite their functional diversity, comparison of TMDs for the four Type I importers revealed that the four helices that display “up and down” topology form a common arrangement of TM fold for Type I importers.
Figure 1. Crystal structures of Type I ABC importers. The TMD subunits are colored pink and deep purple, the NBD subunits are colored light cyan and teal, SBPs orange, RDs dark and light green, the regulation factor EIIA is pale yellow, the substrates are blue spheres and nucleotides are red spheres. From left to right on the top row, the following structures are represented in nucleotide-free conformations: ModBC of A. fulgidus [AfModB2C2-A], ModBC of M. acetivorans [MaModB2C2], and methionine transporter MetN2I2 from E. coli. MaModB2C2 is shown in the inhibited state with molybdate bound to the RDs. The bottom row shows structures of the maltose transporter from E. coli, MalFGK2-E in the following conformations (from left to right): inward-facing; pre-translocation state; outward-facing, nucleotide bound; and inhibited, EIIAglu-bound.
![Figure 1. Crystal structures of Type I ABC importers. The TMD subunits are colored pink and deep purple, the NBD subunits are colored light cyan and teal, SBPs orange, RDs dark and light green, the regulation factor EIIA is pale yellow, the substrates are blue spheres and nucleotides are red spheres. From left to right on the top row, the following structures are represented in nucleotide-free conformations: ModBC of A. fulgidus [AfModB2C2-A], ModBC of M. acetivorans [MaModB2C2], and methionine transporter MetN2I2 from E. coli. MaModB2C2 is shown in the inhibited state with molybdate bound to the RDs. The bottom row shows structures of the maltose transporter from E. coli, MalFGK2-E in the following conformations (from left to right): inward-facing; pre-translocation state; outward-facing, nucleotide bound; and inhibited, EIIAglu-bound.](/cms/asset/3a3ad00d-5bb4-4a10-aec7-85c832951828/ibmg_a_953626_f0001_b.jpg)
In addition, some Type I importers posses a regulatory domain (RD) attached to the highly conserved NBDs, which controls the function of these importers. Three of the four structurally characterized Type I importers (all except ModBC of A. fulgidus [AfModBC]; Hollenstein et al., Citation2007) have RDs. The RDs of MalFGK (Oldham et al., Citation2007) and ModBC of M. acetivorans [MaModBC] (Gerber et al., Citation2008) have a similar overall fold displaying an elongated beta-barrel composed of two beta-sandwiches. The RD of MetNI exhibits a ferredoxin-like fold (Kadaba et al., Citation2008). Once substrate or regulatory proteins bind to the RD, ATP hydrolysis and substrate transport are halted. By modulating transport activity, accessory domains such as the RDs can provide additional function to the core transporter.
Type I importers transport substrate by an alternating access mechanism
Although the number of TMD helices in Type I importers can vary, they transport their substrates by the same alternating access model (Jardetzky, Citation1966). Importers alternate between two conformations, inward- and outward-facing where the translocation pathway will be exposed to only one side of the membrane.
The detailed transport mechanism employed by Type I transporters can be explained by the maltose transporter, which is one of the best-characterized Type I systems. The transport of maltose is initiated by the binding of substrate-loaded MalE to nucleotide-free MalFGK (Oldham & Chen, Citation2011; , State 1). Substrate-loaded MalE triggers a conformational change of MalFGK, inducing partial closure of the MalK dimer (, State 2). When the NBDs are in the nucleotide-free semi-open state and substrate-loaded MalE is bound, the TMD subunits rotate toward the center of the transporter, narrowing the translocation pathway. However, the periplasmic side of the transporter still remains closed. The binding of two ATP molecules to MalFGK in the pre-translocation state induces conformational changes, including closure of the MalK dimer, opening of MalE, and a shift to the outward-facing conformation of the TMDs by a rigid-body rotation (Oldham et al., Citation2007; , State 3). EPR studies show that MalE alternates between an open and closed conformation depending on the binding of ATP to MalK (Austermuhle et al., Citation2004). Taken together with the result that both MalE and ATP are required to close the MalK dimer (Orelle et al., Citation2008), the EPR studies indicate that communication between MalE and MalK across the membrane is a major step of the transport process. By shifting from the pre-translocation state to the outward-facing state, MalFGK is able to transfer maltose from MalE to a temporary binding site in MalF (Oldham et al., Citation2013; , State 4). Upon hydrolysis of ATP, ADP and inorganic phosphate are released and MalFGK returns to the resting, inward-facing conformation. In this state, the closed MalK dimer is opened and MalE departs from the transporter.
Figure 2. Mechanism of Type I ABC importers. Two TMD subunits are colored purple and pink, and two NBD subunits are colored as cyan and magenta. The periplasmic binding proteins are colored orange and the substrate is represented by a blue sphere. ATP and ADP are shown as a red and yellow sphere, respectively. The substrate-loaded SBP binds to inward-facing conformation of transporter (State 1), and induces a partial closure of two NBD subunits (State 2). Binding of ATP triggers closure of the two NBDs, reorientation of TMD subunits, and opening of the SBP, resulting in release of substrate to the TMD subunits (State 3). The dissociation of ADP molecules resets the transporter back to an inward resting state (State 4). The inhibited state (State 5) is shown in brackets; when cytoplasmic substrate levels are high enough, substrate binds the RDs preventing conformation change and ATP hydrolysis of the transporter.
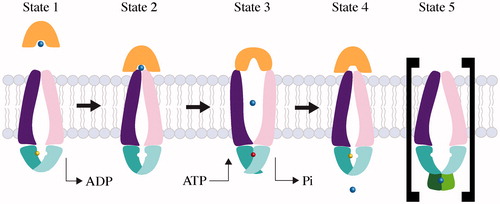
Additional structures of Type I ABC importers have confirmed the mechanism defined by the maltose transporter. The structures of ModBC from A. fulgidus [AfModBC] (Hollenstein et al., Citation2007), and MetNI (Kadaba et al., Citation2008) further confirm the conformation of the resting state for a Type I transporter; without nucleotide and in complex with substrate-loaded SBP. In the resting state, Type I importers adopt an inward-facing conformation, where the translocation pathway is open to the cytoplasm and the two NBDs adopt nucleotide-free, open conformation. Then, binding of ATP to the NBD dimer brings the NBD subunits together as observed in nucleotide-bound MalFGK (Oldham et al., Citation2007) resulting in outward-facing conformation where the substrate-binding site will be exposed to the periplasm.
Due the presence of the regulatory domain, an additional conformation is allowed in Type I importers. The structure of MaModBC represents the trans-inhibited state, where two tungstate ions were bound to the regulatory domains of the ModC NBD dimer. (, State 5; Gerber et al., Citation2008). In this trans-inhibited state, MaModBC displays a larger distance between the coupling helices relative to other importers. The bound tungstate ions inhibit transport in MaModBC by preventing the closure of the NBD dimer around ATP. In fact, the only contacts between the two ModC subunits in the trans-inhibited state are formed through the regulatory domains.
The Type I importer structures determined to date have a general theme; open NBDs are coupled with inward-facing conformation of importers and outward-facing conformation of importers will be induced by binding of ATP to the NBDs resulting in closed NBD dimer. Alternation between these two conformations allows movement of substrate across the membrane.
Initiation of the transport cycle is induced by substrate-binding protein
The transport cycle is initiated by the interaction of the SBP with the TMDs. While SBPs were originally divided into three classes according to the arrangement of the core beta-sheets (Fukami-Kobayashi et al., Citation1999), a growing amount of data lead to a reclassification of SBPs into six clusters based on their three-dimensional structures (Berntsson et al., Citation2010). Notably, the substrate-binding proteins of structurally characterized Type I importers belong to Clusters B and F (Berntsson et al., Citation2010). However, predicted Type I transporters such as OppBCDF or PotBCD share operons with SBPs of other clusters including C and D, respectively (Berntsson et al., Citation2010; Cserzo et al., Citation2002). It is suggested that most of SBPs consists of two globular domains that are separated by a hinge wherein the ligand is bound and enclosed by an inter-domain rotation (Duan et al., Citation2001; Hollenstein et al., Citation2007; Spurlino et al., Citation1991). Ligand binding brings about a large conformational change in the protein (Sharff et al., Citation1992; Shilton et al., Citation1996), and this ligand-induced conformational change appears to be a general feature of the SBPs known to associate with Type I importers. Both the loaded and unloaded forms of MalE can bind to MalFGK (Austermuhle et al., Citation2004; Davidson et al., Citation1992). However, only binding of substrate-loaded MalE to transporter can induce the complete closure of two NBDs.
Additional TMD loop contributes to the unique transport process in the maltose transporter
In the maltose transporter, the interaction between MalE and the periplasmic MalF-P2 loop mediates substrate recognition by inducing an activated conformational change of MalE, which precedes substrate transport (Jacso et al., Citation2012). The P2 loop is a large Ig-like periplasmic domain (P2 loop ∼20 kDa) connecting TM helices 3 and 4 and is only observed in the MalF homologues of enteric bacteria (Ehrmann et al., Citation1998). The P2 loop raps around MalE, forming a receptor that recognizes the SBP and allows MalE and MalF to remain in close contact throughout the catalytic cycle (Daus et al., Citation2009; Jacso et al., Citation2009). A structural comparison of P2 loop from the two different states of maltose transporter (Oldham & Chen, Citation2011; Oldham et al., Citation2007) indicated that the P2 loop is highly flexible and plays a key role to maintain contacts with MalE regardless of the conformational state of transporter. The flexibility of the P2 loop was further supported by structure of maltose transporter in inward-facing state where flexible P2 loop was not observed in the absence of MalE (Khare et al., Citation2009).
Post-translational regulation of Type I importers is dependent on the regulatory domain
The regulation mechanism employed by Type I importers is distinct for each transporter, even if they share a similar RD fold such as MalFGK and MaModBC or transport the same substrate such as AfModBC and MaModBC.
The RD of the maltose transporter contributes to regulatory functions by binding MalT, a transcription factor that is part of the maltose operon, or IIAglc , an enzyme that inhibits transport activity (Boos & Shuman, Citation1998). Recently, the complex structure of MalFGK with IIAglc revealed that two IIAglc bind the MalK dimer (Chen et al., Citation2013). This interaction inhibits transport by preventing the closure of the MalK dimer and stabilizing MalFGK in the inward-facing resting state (similar to , State 5).
The regulation of MaModBC (Gerber et al., Citation2008) and MetNI (Kadaba et al., Citation2008) was accomplished by binding of substrates to the structurally distinct RDs likewise inhibiting closure of the NBD dimer. This indicated that MetNI was regulated through trans-inhibition similar to MaModBC, a conclusion supported by selenomethionine soaking which identified a substrate-binding site in MetN RD (Kadaba et al., Citation2008). There are two mechanisms known to regulate ModBC. Binding of molybdate directly to the RD as in the MaModBC and repression of modABC expression by molybdate-dependent transcription regulator ModE. The substrate-binding C-terminal domain of ModE has similar overall fold as the RD of MaModBC. ModE changes its conformation by binding molybdate (Hall et al., Citation1999; Schuttelkopf et al., Citation2003), which allows the N-terminal DNA binding domain of ModE to act as a repressor of the molybdate transporter (Mouncey et al., Citation1996). This indicates that Type I transporters without RDs like AfModBC (Hollenstein et al., Citation2007) can be regulated by alternative mechanisms (Grunden et al., Citation1996).
Type II ABC importers
There are several characteristics that distinguish Type II transporters from Type I. The association of SBP with the Type II transporters is generally weakened by the presence of substrate (Lewinson et al., Citation2010; Vigonsky et al., Citation2013). Type II transporters have a greater number of TM helices (Locher, Citation2009; Oldham et al., Citation2008), with the core helices undergoing distinct conformation changes to transport substrate (Korkhov et al., Citation2012b; Rice et al., Citation2013). While Type II transporters share these overall similarities, there is mounting evidence to suggest mechanistic differences within the Type II sub-family (Rice et al., Citation2013, Citation2014). With substrates ranging from molybdate to vitamin B12, the variety of substrate size may offer some explanation for the observed diversity among Type II transporters.
SBP-mediated uptake in Type II systems
Similar to the Type I importers, substrate specificity in Type II transport systems is defined by SBPs. Interestingly, despite a wide range in substrate size and chemistry, the structurally characterized Type II importers bind to the same class of SBP (Cluster A, Class III; Borths et al., Citation2002; Mattle et al., Citation2010; Tirado-Lee et al., Citation2011). The distinguishing characteristic of Cluster A SBPs is a rigid α-helix in the hinge region connecting two substrate-binding lobes (Berntsson et al., Citation2010). Other SBPs have a flexible hinge region, and substrate-binding forces the lobes to undergo a “Venus flytrap” conformation change (Mao et al., Citation1982). Substrate-dependent conformation change in Cluster A SBPs is limited to local loop rearrangement and up to a few degrees of closure (Berntsson et al., Citation2010; Karpowich et al., Citation2003). Even though Cluster A SBPs bind substrates ranging over a 10-fold change in volume, the majority of structural differences between the different SBPs are limited to the loops in the substrate-binding pocket (Tirado-Lee et al., Citation2011).
With only three Type II transporters structurally characterized and limited biochemical data on Type II complexes, it is possible that other types of binding protein may bind with Type II transporters. For example, the Cluster B L-arabinose binding protein ABP (E. coli gene name araF; Quiocho et al., Citation1974) shares an operon with the putative Type II transporter AraHGp (Scripture et al., Citation1987); AraHp is predicted to have 10 helices in each of the TMDs. Likewise in Pasteurella haemolytica, the Cluster D SBP FbpA (Shouldice et al., Citation2004) shares an operon with the putative Type II transporter FbpBCp (Kirby et al., Citation1998), which is predicted to have an 11 TM helix topology. Although the complex formation of ABP to AraHGp or FbpA to FbpBCp has yet to be tested, these cases may broaden the diversity of SBPs in Type II transporter complexes.
Despite limited effects of substrate on SBP conformation in Type II systems, substrates significantly alter the affinity of SBP for Type II transporters (Lewinson et al., Citation2010; Vigonsky et al., Citation2013). In the absence of substrate, Type II transporters form high affinity complexes with their cognate binding proteins. Substrate diminishes this affinity either by increasing the dissociation rate (as for BtuCD; Lewinson et al., Citation2010) or by decreasing the association rate (as for MolBC; Vigonsky et al., Citation2013). The effects of ligand on Type II complex formation are inverted in Type I transporters, which require nucleotide or substrate for complex formation (Vigonsky et al., Citation2013). A close comparison of the affinities of MolA for molybdate and MolBC for MolA has suggested that molybdate weakens complex association through an allosteric binding site on MolBC (Vigonsky et al., Citation2013). However, a substrate-binding site has yet to be observed for a Type II transporter, though such sites have been reported in Type I transporters (Gerber et al., Citation2008; Oldham et al., Citation2007).
Structural analysis elucidates the diversity and conformational changes in Type II transporters
The Type I and II importers were first segregated based on differences in the size and overall architecture of the core transporters (Locher, Citation2009; Oldham et al., Citation2008). Type II transporters are larger, with the structurally characterized TMD homodimers having 20 TM helices (10 helices per monomer identified as TM 1–10). Close comparison of the TMDs has suggested that the majority of conformation changes are limited to the core helices TM 3, 4, 5, 5a, 10 and the loop between TM 2 and 3. This is distinct from Type I transporters, which undergo domain-wide rigid-body conformation changes as described in the above section.
The core helices control access to the translocation pathway and are central in the transport mechanism. The three structurally characterized Type II core transporters are shown in with their cognate SBPs poised on the periplasmic side of the TMDs. The translocation pathway has been highlighted using MolAxis (Yaffe et al., Citation2008), which emphasizes the different conformations of each structure and the accessibility of the translocation pathway. The nucleotide-free conformations of BtuCD and HmuUV are similar showing a central translocation pathway at the TMD interface lined by TM5, 5a, 10 and the TM2-3 loop (Locher et al., Citation2002; Woo et al., Citation2012). In these apo structures, the translocation pathway is in an outward-facing conformation with an occlusion formed by the cytoplasmic side of TM5 (cytoplamic gate I). The key difference between the two transporters is the volume of the translocation pathway. With an internal volume of ∼3400 Å3, BtuCD is able to accommodate vitamin B12 with few steric clashes, while HmuUV with an internal cavity of ∼1300 Å3 is able to accommodate one molecule of heme (cavity volumes calculated with 3VEE web server and a large and small probe radius of 10 and 2 Å respectively (Voss & Gerstein, Citation2010)).
Figure 3. Conformational diversity among Type II importer crystal structures with MolAxis prediction of the translocation pathways. The TMD subunits of each transporter are shown as dark purple and pink tubes, the NBD subunits are teal and cyan, the SBPs are orange, and substrates are light blue. The MolAxis program was used to find and analyze the translocation pathway by fitting the largest possible spheres in along the central cavity (Yaffe et al., Citation2008). Spheres with a diameter <1.4 Å are colored red, 1.4–4 Å green and >4 Å blue. (A) Type II core importers MolB2C2 from H. influenza, HmuU2V2 from Y. pestis and BtuC2D2 from E. coli each solved in the absence of nucleotide. Cognate SBPs (MolA, HmuT and BtuF, respectively) are shown poised above the periplasmic gates. (B) Type II importers solved in complex with SBP. The BtuCD-F structure on the right was solved with AMP-PNP bound (nucleotide not shown).
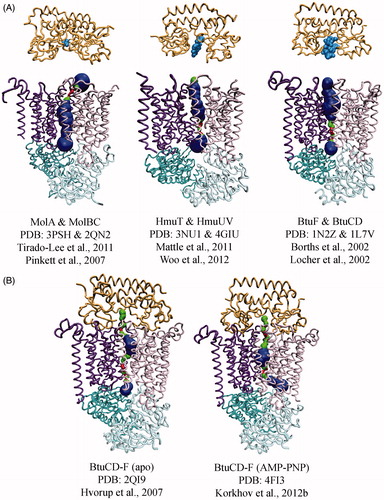
A greater conformational difference is observed with nucleotide-free MolBC, where a closed periplasmic gate (formed by TM 5a) and the open cytoplasmic gate I positions the transporter in the inward-facing conformation (Pinkett et al., Citation2007). It is notable that access through the periplasmic gate appears to correlate with the size of substrate, suggesting that the translocation pathway may add to substrate selectivity with the same filter mechanism as a channel.
BtuCD has also been solved in complex with its SBP BtuF in the presence (Korkhov et al., Citation2012b) and absence (Hvorup et al., Citation2007) of nucleotide (), which provides us with information on the conformational flexibility of a Type II importer. The nucleotide-free BtuCD-F complex displayed an asymmetric conformation of TM5 and 5a, with one TMD subunit resembling the outward-facing, un-complexed BtuCD and the other subunit resembling the inward-facing MolBC (Hvorup et al., Citation2007). This conformation of the translocation pathway was occluded, leaving inadequate space for substrate. In the presence of nucleotide, a large internal cavity was formed between a closed periplasmic gate and a closed second cytoplasmic gate (gate II) formed by the TM2–3 loop. Although, substrate was not present in the structure, the cavity is large enough to hold vitamin B12 with few steric clashes (Korkhov et al., Citation2012b).
Mechanistic details of the Type II importer, BtuCD
Structural snap-shots, biochemical analysis and electron paramagnetic resonance (EPR) spectroscopy of BtuCD-F have culminated in a well-defined Type II transport mechanism. Here, the mechanism can be described as four states. Analysis of complex association rates in an excess of ATP-Mg2+ has suggested that the majority of a BtuCD transporter population can be found in the ADP-bound state (, State 1; commonly known as the post-hydrolysis state; Lewinson et al., Citation2010), which has a nearly identical conformation to the nucleotide-free transporter (Joseph et al., Citation2011, Citation2014). With the periplasmic gate (TM 5 and 5a) open and cytoplasmic gate I (TM5) closed, the nucleotide-free state is outward-facing (Locher et al., Citation2002). Substrate-loaded BtuF binds to BtuCD, delivering substrate to the outward-facing translocation pathway (, State 2; Borths et al., Citation2005; Locher et al., Citation2002).
Figure 4. Mechanisms of large and small substrate Type II transporters. The TMD subunits are colored dark purple and pink, the NBD subunits are teal and cyan, the SBP is orange and substrates are light blue. The colors of TMD, NBD, SBP and substrate are the same as in . (A) Transport of a large substrate is initiated when substrate-loaded binding protein docks to the periplasmic surface of the transporter. In State 1, the transporter is shown with ADP (yellow) bound, but the nucleotide-free state is conformationally identical. In State 2, we show SBP docking before ATP due to the higher affinity of transporter for loaded SBP in the absence of nucleotide (Lewinson et al., Citation2010) and the slightly more open conformation of the BtuCD-F periplasmic gate in the absence of nucleotide (Joseph et al., Citation2011). State 3 shows that the binding of ATP (red) traps substrate (blue) in a translocation chamber. Post-hydrolysis, the NBDs open allowing the departure of phosphate and the concurrent collapse of the translocation chamber. Substrate is depicted as it is being squeezed into cytoplasm by the transporter progressing toward an occluded complex (State 4). The transport cycle restarts when the occluded complex disassembles. (B) Transport of a small substrate is initiated when loaded SBP binds to the transporter (State 1). Though small substrate has yet to be trapped in a Type II complex, we suspect that substrate cannot be transferred until ATP binds and the periplasmic gate opens (State 2). After transfer, small substrate may be held in an internal cavity comparable to large substrates (State 3). The possibility of reversion from State 3 to State 2 cannot be ruled out, and may be distinct from the sealed cavity of large substrate Type II transporters. After nucleotide is hydrolyzed, the small substrate transporter returns to an apo-like inward-facing state, from which substrate can enter the cytoplasm (State 4).
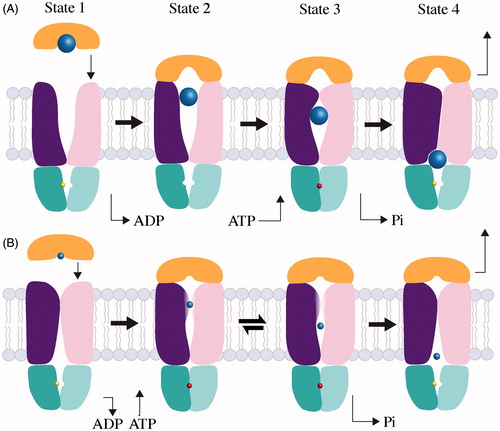
The BtuCD-F complex has very little affinity for vitamin B12 (Borths et al., Citation2005; Lewinson et al., Citation2010), and substrate transfer may be encouraged by a slight opening of the BtuF lobes and loops from the periplasmic gate reaching toward the substrate-binding pocket (Hvorup et al., Citation2007). The ADP bound transporter, with a closed cytoplasmic gate I, prevents passage of substrate to the cytoplasm (Goetz et al., Citation2009; Joseph et al., Citation2011). When BtuCD-F binds nucleotide (, State 3), the NBDs close, pulling the coupling helices closer together. This triggers the closing of the periplasmic gate and concomitantly, the opening of cytoplasmic gate I and closing of cytoplasmic gate II, which traps substrate in a large translocation chamber (Korkhov et al., Citation2012b; Joseph et al., Citation2011, Citation2014).
Studies with ATP analogs representing the pre-hydrolysis ATP bound state (ATP + EDTA, AMP–PNP) and the intermediate state before phosphate departure (ADP + Mg2+ + vanadate) show similar conformations. This suggests that post-hydrolysis conformation changes coincide with phosphate departure (Joseph et al., Citation2014). When nucleotide is hydrolyzed, the NBDs separate, phosphate departs, cytoplasmic gate II opens and the translocation chamber collapses with the TM5 helices squeezing vitamin B12 into the cytoplasm (Joseph et al., Citation2014). After the departure of substrate, BtuCD-F is in a stable, occluded conformation (, State 4; Hvorup et al., Citation2007, Korkhov et al., Citation2012a). The mechanism for disassociation of the Type II transporter complex is not well understood, but Lewinson et al. (Citation2010) have suggested that basal ATP hydrolysis and possibly free substrate may play important roles (Vigonsky et al., Citation2013).
Mechanistic diversity among Type II transport systems
Studies of the molybdate importer MolBC have uncovered novel characteristics of the Type II transporter mechanism. EPR and cross-linking studies of MolBC-A confirmed the nucleotide-free, inward-facing crystal structure of MolBC (Pinkett et al., Citation2007; Rice et al., Citation2013, Citation2014; , State 1). In MolBC, the nucleotide-dependent movement of the periplasmic gate is more limited than BtuCD, with TM 5a shifting apart enough to disrupt disulfide cross-linking. This increases the mobility of spin-labels along the helix and presumably allows the passage of a relatively small substrate like molybdate (Rice et al., Citation2013). In the presence of nucleotide, we suspect that substrate passes from the SBP into the translocation pathway (, State 2; Rice et al., Citation2013). When nucleotide is bound to the transporter, the cytoplasmic ends of the TM 5 helices shift away from each other and do not assume a closed conformation, and thus this region of MolBC cannot be considered a gate (Rice et al., Citation2013). The outward movement of TM 5 is likely coupled to the inward movement of cytoplasmic gate II, which closes in the presence of nucleotide. The resulting conformation (, State 3) is predicted to be similar to that of BtuCD-F (, State 3), with substrate trapped in the translocation chamber. (Rice et al., Citation2013, Citation2014). For MolBC-A, there is currently no evidence to support an occluded conformation. Instead hydrolysis of ATP returns the transporter to an apo-like inward-facing state (, State 4), where substrate is allowed to enter the cytoplasm (Rice et al., Citation2013). It is possible that the mechanistic differences between MolBC and BtuCD can be traced back to the size of substrate, where a large opening of the periplasmic gate was not required for molybdate passage. And, with the periplasmic gate closed in the nucleotide-free conformation, cytoplasmic gate I was not required (Rice et al., Citation2013). It remains to be seen if the MolBC mechanism will be observed for other small-substrate Type II importers or if additional variations on the Type II transporter mechanism will be found.
Given the differences in substrates, it should come as no surprise that the structures and mechanisms of Type II transporters have diversified. As we continue to explore the Type II subfamily, we can expect to find additional sources of diversity. For Type II transporters, the biggest questions remain in the area of regulation. Whereas Type I transporters have well-described mechanisms of transcriptional and post-translational regulation, regulation of Type II transport is largely unexplored. Like Type I transporters, Type II systems can be regulated by substrate at the transcription level [MolBCA (Rice et al., Citation2014), HmuUVT (Chao et al., Citation2005) and FhuBCD (Andrews et al., Citation2003)]. A mechanism of post-translational regulation for a Type II system has yet to be verified, but a thorough database search (http://exon.niaid.nih.gov/abctransporter/) can reveal extensions to the core transporter that may prove to be Type I-like RDs or other accessory domains.
The emergence of ECF transport system: Type III transporters?
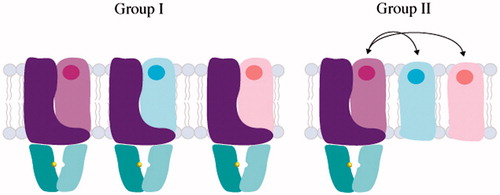
Energy-coupling factor (ECF) transporters were first discovered in the 1970s (Henderson et al., Citation1979), but only recently classified as a new type of ABC transporter (Rodionov et al., Citation2009). These proteins play a critical role in micronutrient uptake in bacteria and archaea. Similar to Type I and II transporters, ECF transporters consist of two nucleotide-binding domains (EcfA and EcfA′), a transmembrane coupling domain (EcfT) and a substrate-binding component (EcfS). EcfA components can assemble as homodimers or heterodimers and the EcfA–EcfA′–EcfT complex forms the energizing module. While Type I and II ABC importers have a substrate-binding protein to deliver compounds to the translocation pathway, ECF transporters contain a unique feature: the EcfS component. The EcfS, or S component, is an integral membrane protein that binds substrate with nanomolar affinity (Duurkens et al., Citation2007; Hebbeln et al., Citation2007). S components also differ in their genetic organization and the way they interact with the energy module, separating ECF transporters into two subgroups (). For Group I ECF transporters, each S component forms a complex with a unique energizing module and the S-components are located within the same operon. Alternatively, Group II ECF transporters consist of modular S components that bind to a diverse array of substrates, yet multiple S components share the same energizing module to make up the full transporter (S, T, A and A′). For Group II ECF transporters, the S components are scattered across the genome. All three energizing components (T, A, A′) are necessary for the S component to form a complex, and all four components (T, S, A and A′) of the ECF transporter are necessary for transport of substrate across the membrane (Karpowich & Wang, Citation2013). Despite the diversity of substrates and assembly that differentiate these transporters from Type I and II importers, ECF transporters also use the energy of ATP to translocate substrates across the lipid bilayer.
Figure 5. Domain architecture and assembly of Group I and II ECF transporters. ECF transporters contain an energy module (A and A′, colored in teal and cyan respectively), transmembrane component (T, colored in purple) and a substrate-binding module (S, colored in light purple, light blue and light pink). In Group I ECF transporters, S associates with dedicated T, A, A′ units, while Group II ECF transporters utilize the same T, A and A′ components for several different S components.
The S component is responsible for substrate binding
From thiamine to riboflavin, S components bind a diverse range of substrates. To date, structures of the S components ThiT (thiamine; Erkens et al., Citation2011) and BioY (Biotin) from Lactococcus lactis (Berntsson et al., Citation2012), RibU (riboflavin) from Staphyloccus aureus (Zhang et al., Citation2010) and NikM (nickel) from Thermoanaerobacter tengogensis (Yu et al., Citation2014) have been solved (). In addition, the complete complex structures of the Folate (Xu et al., Citation2013) and hydroxymethyl pyrimidine (HMP) (Wang et al., Citation2013) transporters from Lactococcus brevis were solved in the absence of substrate ().
While not sequentially similar, the structures of RibU, BioY and ThiT reveal that helices 4, 5 and 6 directly interact with the riboflavin, biotin and thiamine, respectively. The long L1 loop between helices 1 and 2 covers the substrate-binding pocket in the substrate bound structures, but in the full length structure, this loop does not cap the binding pocket, potentially indicating a substrate sensing mechanism. Understandably, there is variation in sequence amongst the EcfS binding pocket that accounts for specificity, but the S components have substrate affinity in the nM to pM range. NikM, the S component of the ttNikM2ECF transporter from Thermoanaerobacter tengcogensis (Yu et al., Citation2014) fits into a subclass of S component metal transporters that are part of the Group I transport system. Similar to the nutrient EcfS components, NikM contains the core 6 helices and has two additional modifications that make this group unique. The first nine residues that make up a loop proceeding helix 1 are conserved across ECF metal S components. This N-terminal loop would block the vitamin binding sites found in other S components. In combination with an additional N-terminal helix, both form a binding site specific for metals that would prevent vitamin binding (Yu et al., Citation2014).
Full-length structures shed light on assembly and mechanism of ECF transporters
The most recent structures of the Folate and HMP ECF transporter from L. brevis ( Wang et al., Citation2013; Xu et al., Citation2013) have provided us with a glimpse into the assembly and mechanism of this class of transporters. These full structures provide the first view of the EcfT domain and how the ECF components assemble to make an intact ECF transporter ().
Figure 6. Crystal structures of Type III ECF importers. (A) Cartoon diagrams of S component crystal structures RibU from S. aureus, ThiT and BioY from L. lactis and NikM2 from T. tengcongensis. Each S component is bound to substrate shown in blue stick representation. (B) Crystal structures of full-length, substrate-free folate and HMP transporters from L. brevis. For both full-length structures, the energizing component (shown in purple) predominantly interacts with the substrate bound component in pink. The EcfA and EcfA′ are shown in cyan and teal.
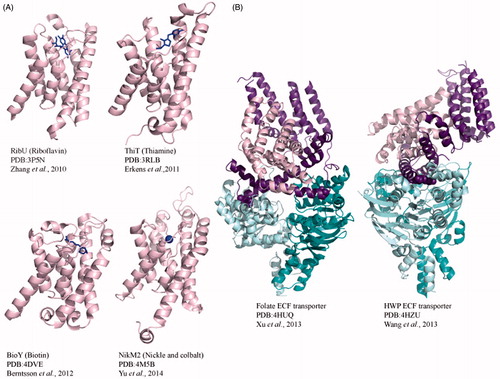
The EcfT domain consists of eight helices, five of which make up the EcfT core domain perpendicular to the membrane and additional helices dipping into the cytoplasmic space, which form an “L” shape configuration. The full-length structures in the absence of substrate reveal how the S component interacts with the energizing component EcfT. The S component fits between the five core helices and cytoplasmic helices, with five of the six EcfS helices making contacts with EcfT. In complex, helices 1, 2 and 6 of EcfS are responsible for most of the hydrophobic contacts with EcfT. The structures are supported by studies on the conserved motif (AXXXA, X mostly hydrophobic) in helix 1 shown to be essential for interactions with the ECF energizing module (Berntsson et al., Citation2012; Erkens et al., Citation2011).
Structurally different from the configuration of Type I and II transporters, the cytoplasmic EcfT helices (but not the EcfS helices) make direct contacts with EcfA and EcfA′. EcfT helices 6 and 7 both contain a conserved sequence called the coupling module, XRX, which directly interacts with a groove in each of the nucleotide binding domains. Double mutations of the conserved arginine residues result in a dissociation of the ECF complex and loss of ATPase activity (Neubauer et al., Citation2009). Coupling helices are also present in ABC importers and exporters, as a means of communication between the TMD and NBD (Locher, Citation2009).
The dimerized structure of the EcfA–EcfA′ in the full length structures is similar to the structures of homodimer CbiO of CbiMNQO the cobalt EcfA–EcfA′ energy module from Thermoanaerobacter tengcongensis (Chai et al., Citation2013) and EcfA–EcfA′ heterodimer structure from T. maritima (Karpowich & Wang, Citation2013). Similar to Type I and II transporters, the NBD is made up of a helical subdomain and RecA-like domain with the conserved motifs for ATP binding and hydrolysis. The structures of the ECF transporter show two C-terminal helices, which form a four-helix bundle upon dimerization not common to all ABC importers.
Although both full-length ECF structures are substrate and nucleotide free, they start to provide insights into a possible mechanism. The EcfA–EcfA′ conformation is most similar to a nucleotide free open conformation, leading one to believe that the structure is currently in an inward-facing conformation. For both Group I and II ECF transporters, it is assumed that ATP induced conformational changes are coupled to the substrates translocation and subsequent release into the cytoplasm. The structures reveal that the S component of the ECF transporter lies parallel to the membrane surface, presumably in a conformation to allow for substrate release into the cytoplasm. In both full-length structures, the angle of the S component makes the opening of the substrate-binding pocket accessible to the lipid–cytoplasm interface. Based on this conformation, the proposed mechanism would begin with the nucleotide free resting state, where the EcfS component is perpendicular to the EcfT component in the membrane and the EcfA–EcfA′ components are far apart. ATP binding would bring EcfA–EcfA′ closer together into a closed conformation and rotate the EcfS module into a parallel position so it could bind extra-cellular substrate. ATP hydrolysis would rotate the ECF transporter back to the resting state for subsequent substrate release in the cytoplasm. As we go forward with these Type III ABC importers, the major questions will include connecting substrate translocation to nucleotide states and the conformational changes necessary to trigger the release of substrate. We await the substrate-loaded and nucleotide-bound full-length structures of ECF transporters to shed light on the proposed mechanisms.
Conclusion
The culmination of genetic, structural and biochemical studies of ABC importers to date have revealed three different systems that utilize ATP to transport an array of substrates across cellular membranes. While the overall architectural differences between Type I, II and III transporters reflect diversity in both the mechanism of transport and regulation, these systems only scratch the surface on the complexity of this family. ABC importers have evolved to use multiple mechanisms not only to transport nutrients across the membrane but also to survive in an ever-changing and hostile environment (Kjelleberg et al., Citation1993; Lewis et al., Citation2012; Lin et al., Citation2009).
Genome analysis uncovered multiple importers that transport the same substrate within an organism. Studies of the mol and mod systems in H. influenzae revealed the co-existence of a low and high affinity transport system for molybdate in the same organism (Rice et al., Citation2014). This duplication is also seen in E. coli, albeit with less specificity for one substrate. Here, cys, mod and an additional nonspecific transport system are responsible for molybdate and sulfate transport (Aguilar-Barajas et al., Citation2011; Sirko et al., Citation1995). In several bacterial organisms, the presence of two sulfate transport systems are found in separate operons, suggesting an importance of redundancy in uptake systems for particular bacteria (Aguilar-Barajas et al., Citation2011).
We are also finding unexpected diversity in the substrates bound by particular SBPs and transport systems. The sap operon (sapABCDFZ) from non-typable Haemophilus influenzae (NTHI) encodes for importers that select for antimicrobial peptides (AMPs) for transport into the cytoplasm and subsequent degradation, presumably before peptide accumulation can cause irreparable damage to the cytoplasmic membrane. The SapA binding protein has the ability to select for both heme and AMPs, present in different microenvironments in the host (Mason et al., Citation2011; Shelton et al., Citation2011). This mechanism also seems to be conserved in other bacteria, which need to survive under different environmental stresses (Tanabe et al., Citation2007; Vanderlinde et al., Citation2010). These systems not only suggest diversity in substrate recognition but also possible variation in the complexes formed between SBP and transmembrane domains.
While it was once thought that ABC importers utilized a single SBP per transporter located on the same operon, we now see examples of multiple SBPs per transporter complex. For example, GlnPQ importer from E. faecalis and S. pneumonia has multiple SBPs with high sequence similarity, yet each differ in their affinity to glutamine. In addition these SBPs can also bind asparagine with low affinity (Fulyani et al., Citation2013), reflecting the diversity introduced through the SBPs that interact with the same transport system.
As we begin to structurally and functionally elucidate additional transport systems with an overall architecture that differs from what is currently known, there is the potential to discover an even greater diversity among these complex systems. Going forward, our comprehension of transport systems will depend not only on understanding the details of what distinguishes a Type I importer from a Type II or the role of size of substrate in the mechanism of transport, but also an overall analysis of the transport systems in a given organism. The multiplicity in transport components, whether it is duplicate transporters or multiple SBPs that interact with the same transporter make these systems multifunctional. Since nutrient uptake is interconnected to bacterial survival and pathogenesis, a detailed understanding of the interplay between transport systems will be essential.
Declaration of interest
The authors report no declarations of interest. This work is funded by NSF grant MCB1121872.
Acknowledgements
We thank Kari Tanaka for critical insight and discussion on the manuscript.
References
- Aguilar-Barajas E, Diaz-Perez C, Ramirez-Diaz MI, et al. (2011). Bacterial transport of sulfate, molybdate, and related oxyanions. Biometals 24:687–707
- Al-Shawi MK. (2011). Catalytic and transport cycles of ABC exporters. Essays Biochem 50:63–83
- Andrews SC, Robinson AK, Rodriguez-Quinones F. (2003). Bacterial iron homeostasis. FEMS Microbiol Rev 27:215–37
- Austermuhle MI, Hall JA, Klug CS, Davidson AL. (2004). Maltose-binding protein is open in the catalytic transition state for ATP hydrolysis during maltose transport. J Biol Chem 279:28243–50
- Bao H, Duong F. (2012). Discovery of an auto-regulation mechanism for the maltose ABC transporter MalFGK2. PLoS One 7:e34836
- Berntsson RP, Smits SH, Schmitt L, et al. (2010). A structural classification of substrate-binding proteins. FEBS Lett 584:2606–17
- Berntsson RP, Ter Beek J, Majsnerowska M, et al. (2012). Structural divergence of paralogous S components from ECF-type ABC transporters. Proc Natl Acad Sci USA 109:13990–5
- Biemans-Oldehinkel E, Doeven MK, Poolman B. (2006). ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett 580:1023–35
- Boos W, Shuman H. (1998). Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol Mol Biol Rev 62:204–29
- Borths EL, Locher KP, Lee AT, Rees DC. (2002). The structure of Escherichia coli BtuF and binding to its cognate ATP binding cassette transporter. Proc Natl Acad Sci USA 99:16642–7
- Borths EL, Poolman B, Hvorup RN, et al. (2005). In vitro functional characterization of BtuCD-F, the Escherichia coli ABC transporter for vitamin B12 uptake. Biochemistry 44:16301–9
- Chai C, Yu Y, Zhuo W, et al. (2013). Structural basis for a homodimeric ATPase subunit of an ECF transporter. Protein Cell 4:793–801
- Chao TC, Buhrmester J, Hansmeier N, et al. (2005). Role of the regulatory gene rirA in the transcriptional response of Sinorhizobium meliloti to iron limitation. Appl Environ Microbiol 71:5969–82
- Chen SS, Oldham ML, Davidson AL, Chen J. (2013). Carbon catabolite repression of the maltose transporter revealed by X-ray crystallography. Nature 499:364–8
- Cserzo M, Eisenhaber F, Eisenhaber B, Simon I. (2002). On filtering false positive transmembrane protein predictions. Protein Eng 15:745–52
- Daus ML, Grote M, Schneider E. (2009). The MalF P2 loop of the ATP-binding cassette transporter MalFGK2 from Escherichia coli and Salmonella enterica serovar typhimurium interacts with maltose binding protein (MalE) throughout the catalytic cycle. J Bacteriol 191:754–61
- Davidson AL, Dassa E, Orelle C, Chen J. (2008). Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72:317–64, table of contents
- Davidson AL, Shuman HA, Nikaido H. (1992). Mechanism of maltose transport in Escherichia coli: transmembrane signaling by periplasmic binding proteins. Proc Natl Acad Sci USA 89:2360–4
- Dean M, Hamon Y, Chimini G. (2001). The human ATP-binding cassette (ABC) transporter superfamily. J Lipid Res 42:1007–17
- Duan X, Hall JA, Nikaido H, Quiocho FA. (2001). Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: flexibility of tertiary structure and ligand binding. J Mol Biol 306:1115–26
- Duurkens RH, Tol MB, Geertsma ER, et al. (2007). Flavin binding to the high affinity riboflavin transporter RibU. J Biol Chem 282:10380–6
- Eckford PD, Sharom FJ. (2009). ABC efflux pump-based resistance to chemotherapy drugs. Chem Rev 109:2989–3011
- Ehrmann M, Ehrle R, Hofmann E, et al. (1998). The ABC maltose transporter. Mol Microbiol 29:685–94
- Erkens GB, Berntsson RP, Fulyani F, et al. (2011). The structural basis of modularity in ECF-type ABC transporters. Nat Struct Mol Biol 18:755–60
- Erkens GB, Majsnerowska M, Ter Beek J, Slotboom DJ. (2012). Energy coupling factor-type ABC transporters for vitamin uptake in prokaryotes. Biochemistry 51:4390–6
- Fletcher JI, Haber M, Henderson MJ, Norris MD. (2010). ABC transporters in cancer: more than just drug efflux pumps. Nat Rev Cancer 10:147–56
- Fukami-Kobayashi K, Tateno Y, Nishikawa K. (1999). Domain dislocation:a change of core structure in periplasmic binding proteins in their evolutionary history. J Mol Biol 286:279–90
- Fulyani F, Schuurman-Wolters GK, Zagar AV, et al. (2013). Functional diversity of tandem substrate-binding domains in ABC transporters from pathogenic bacteria. Structure 21:1879–88
- Gerber S, Comellas-Bigler M, Goetz BA, Locher KP. (2008). Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science 321:246–50
- Goetz BA, Perozo E, Locher KP. (2009). Distinct gate conformations of the ABC transporter BtuCD revealed by electron spin resonance spectroscopy and chemical cross-linking. FEBS Lett 583:266–70
- Grunden AM, Ray RM, Rosentel JK, et al. (1996). Repression of the Escherichia coli modABCD (molybdate transport) operon by ModE. J Bacteriol 178:735–44
- Hall DR, Gourley DG, Leonard GA, et al. (1999). The high-resolution crystal structure of the molybdate-dependent transcriptional regulator (ModE) from Escherichia coli: a novel combination of domain folds. EMBO J 18:1435–46
- Hebbeln P, Rodionov DA, Alfandega A, Eitinger T. (2007). Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc Natl Acad Sci USA 104:2909–14
- Henderson GB, Zevely EM, Huennekens FM. (1979). Mechanism of folate transport in Lactobacillus casei: evidence for a component shared with the thiamine and biotin transport systems. J Bacteriol 137:1308–14
- Higgins CF. (1992). ABC transporters:from microorganisms to man. Annu Rev Cell Biol 8:67–113
- Higgins CF, Hiles ID, Salmond GP, et al. (1986). A family of related ATP-binding subunits coupled to many distinct biological processes in bacteria. Nature 323:448–50
- Hollenstein K, Frei DC, Locher KP. (2007). Structure of an ABC transporter in complex with its binding protein. Nature 446:213–16
- Hvorup RN, Goetz BA, Niederer M, et al. (2007). Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF. Science 317:1387–90
- Jacso T, Grote M, Daus ML, et al. (2009). Periplasmic loop P2 of the MalF subunit of the maltose ATP binding cassette transporter is sufficient to bind the maltose binding protein MalE. Biochemistry 48:2216–25
- Jacso T, Schneider E, Rupp B, Reif B. (2012). Substrate transport activation is mediated through second periplasmic loop of transmembrane protein MalF in maltose transport complex of Escherichia coli. J Biol Chem 287:17040–9
- Jardetzky O. (1966). Simple allosteric model for membrane pumps. Nature 211:969–70
- Joseph B, Jeschke G, Goetz BA, et al. (2011). Transmembrane gate movements in the type II ATP-binding cassette (ABC) importer BtuCD-F during nucleotide cycle. J Biol Chem 286:41008–17
- Joseph B, Korkhov VM, Yulikov M, et al. (2014). Conformational cycle of the vitamin B12 ABC importer in liposomes detected by double electron-electron resonance (DEER). J Biol Chem 289:3176–85
- Kadaba NS, Kaiser JT, Johnson E, et al. (2008). The high-affinity E. coli methionine ABC transporter:structure and allosteric regulation. Science 321:250–3
- Karpowich NK, Huang HH, Smith PC, Hunt JF. (2003). Crystal structures of the BtuF periplasmic-binding protein for vitamin B12 suggest a functionally important reduction in protein mobility upon ligand binding. J Biol Chem 278:8429–34
- Karpowich NK, Wang DN. (2013). Assembly and mechanism of a group II ECF transporter. Proc Natl Acad Sci USA 110:2534–9
- Kerr ID, Jones PM, George AM. (2010). Multidrug efflux pumps:the structures of prokaryotic ATP-binding cassette transporter efflux pumps and implications for our understanding of eukaryotic P-glycoproteins and homologues. FEBS J 277:550–63
- Khare D, Oldham ML, Orelle C, et al. (2009). Alternating access in maltose transporter mediated by rigid-body rotations. Mol Cell 33:528–36
- Kirby SD, Lainson FA, Donachie W, et al. (1998). The Pasteurella haemolytica 35 kDa iron-regulated protein is an FbpA homologue. Microbiology 144:3425–36
- Kjelleberg S, Albertson N, Flardh K, et al. (1993). How do non-differentiating bacteria adapt to starvation? Antonie Van Leeuwenhoek 63:333–41
- Korkhov VM, Mireku SA, Hvorup RN, Locher KP. (2012a). Asymmetric states of vitamin B(1)(2) transporter BtuCD are not discriminated by its cognate substrate binding protein BtuF. FEBS Lett 586:972–6
- Korkhov VM, Mireku SA, Locher KP. (2012b). Structure of AMP-PNP-bound vitamin B(12) transporter BtuCD-F. Nature 490:367–72
- Lewinson O, Lee AT, Locher KP, Rees DC. (2010). A distinct mechanism for the ABC transporter BtuCD-BtuF revealed by the dynamics of complex formation. Nat Struct Mol Biol 17:332–8
- Lewis VG, Ween MP, McDevitt CA. (2012). The role of ATP-binding cassette transporters in bacterial pathogenicity. Protoplasma 249:919–42
- Lin AE, Krastel K, Hobb RI, et al. (2009). Atypical roles for Campylobacter jejuni amino acid ATP binding cassette transporter components PaqP and PaqQ in bacterial stress tolerance and pathogen-host cell dynamics. Infect Immun 77:4912–24
- Locher KP. (2009). Structure and mechanism of ATP-binding cassette transporters. Phil Trans R Soc B Biol Sci 364:239–45
- Locher KP, Lee AT, Rees DC. (2002). The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296:1091–8
- Mao B, Pear MR, McCammon JA, Quiocho FA. (1982). Hinge-bending in L-arabinose-binding protein. The “Venus's-flytrap” model. J Biol Chem 257:1131–3
- Mason KM, Raffel FK, Ray WC, Bakaletz LO. (2011). Heme utilization by nontypeable Haemophilus influenzae is essential and dependent on Sap transporter function. J Bacteriol 193:2527–35
- Mattle D, Zeltina A, Woo JS, et al. (2010). Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis. J Mol Biol 404:220–31
- Mouncey NJ, Mitchenall LA, Pau RN. (1996). The modE gene product mediates molybdenum-dependent expression of genes for the high-affinity molybdate transporter and modG in Azotobacter vinelandii. Microbiology 142:1997–2004
- Neubauer O, Alfandega A, Schoknecht J, et al. (2009). Two essential arginine residues in the T components of energy-coupling factor transporters. J Bacteriol 191:6482–8
- Oldham ML, Chen J. (2011). Crystal structure of the maltose transporter in a pretranslocation intermediate state. Science 332:1202–5
- Oldham ML, Chen SS, Chen J. (2013). Structural basis for substrate specificity in the Escherichia coli maltose transport system. Proc Natl Acad Sci USA 110:18132–7
- Oldham ML, Davidson AL, Chen J. (2008). Structural insights into ABC transporter mechanism. Curr Opin Struct Biol 18:726–33
- Oldham ML, Khare D, Quiocho FA, et al. (2007). Crystal structure of a catalytic intermediate of the maltose transporter. Nature 450:515–21
- Orelle C, Ayvaz T, Everly RM, et al. (2008). Both maltose-binding protein and ATP are required for nucleotide-binding domain closure in the intact maltose ABC transporter. Proc Natl Acad Sci USA 105:12837–42
- Pinkett HW, Lee AT, Lum P, et al. (2007). An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science 315:373–7
- Quiocho FA, Phillips GN Jr, Parsons RG, Hogg RW. (1974). Letter: Crystallographic data of an L-arabinose-binding protein from Escherichia coli. J Mol Biol 86:491–3
- Rice AJ, Alvarez FJ, Schultz KM, et al. (2013). EPR spectroscopy of MolB2C2-a reveals mechanism of transport for a bacterial type II molybdate importer. J Biol Chem 288:21228–35
- Rice AJ, Harrison A, Alvarez FJ, et al. (2014). Small substrate transport and mechanism of a molybdate ABC transporter in a lipid environment. J Biol Chem 289:15005–13
- Rodionov DA, Hebbeln P, Eudes A, et al. (2009). A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 191:42–51
- Saurin W, Dassa E. (1994). Sequence relationships between integral inner membrane proteins of binding protein-dependent transport systems: evolution by recurrent gene duplications. Protein Sci 3:325–44
- Saurin W, Koster W, Dassa E. (1994). Bacterial binding protein-dependent permeases:characterization of distinctive signatures for functionally related integral cytoplasmic membrane proteins. Mol Microbiol 12:993–1004
- Schneider E, Hunke S. (1998). ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev 22:1–20
- Schuttelkopf AW, Boxer DH, Hunter WN. (2003). Crystal structure of activated ModE reveals conformational changes involving both oxyanion and DNA-binding domains. J Mol Biol 326:761–7
- Scripture JB, Voelker C, Miller S, et al. (1987). High-affinity L-arabinose transport operon. Nucleotide sequence and analysis of gene products. J Mol Biol 197:37–46
- Seeger MA, Van Veen HW. (2009). Molecular basis of multidrug transport by ABC transporters. Biochim Biophys Acta 1794:725–37
- Sharff AJ, Rodseth LE, Spurlino JC, Quiocho FA. (1992). Crystallographic evidence of a large ligand-induced hinge-twist motion between the two domains of the maltodextrin binding protein involved in active transport and chemotaxis. Biochemistry 31:10657–63
- Shelton CL, Raffel FK, Beatty WL, et al. (2011). Sap transporter mediated import and subsequent degradation of antimicrobial peptides in Haemophilus. PLoS Pathog 7:e1002360
- Shilton BH, Shuman HA, Mowbray SL. (1996). Crystal structures and solution conformations of a dominant-negative mutant of Escherichia coli maltose-binding protein. J Mol Biol 264:364–76
- Shouldice SR, Skene RJ, Dougan DR, et al. (2004). Structural basis for iron binding and release by a novel class of periplasmic iron-binding proteins found in gram-negative pathogens. J Bacteriol 186:3903–10
- Sirko A, Zatyka M, Sadowy E, Hulanicka D. (1995). Sulfate and thiosulfate transport in Escherichia coli K-12: evidence for a functional overlapping of sulfate- and thiosulfate-binding proteins. J Bacteriol 177:4134–6
- Slotboom DJ. (2014). Structural and mechanistic insights into prokaryotic energy-coupling factor transporters. Nat Rev Microbiol 12:79–87
- Spurlino JC, Lu GY, Quiocho FA. (1991). The 2.3-A resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J Biol Chem 266:5202–19
- Tanabe M, Mirza O, Bertrand T, et al. (2007). Structures of OppA and PstS from Yersinia pestis indicate variability of interactions with transmembrane domains. Acta Crystallogr D Biol Crystallogr 63:1185–93
- Tirado-Lee L, Lee A, Rees DC, Pinkett HW. (2011). Classification of a Haemophilus influenzae ABC transporter HI1470/71 through its cognate molybdate periplasmic binding protein, MolA. Structure 19:1701–10
- Vanderlinde EM, Harrison JJ, Muszynski A, et al. (2010). Identification of a novel ABC transporter required for desiccation tolerance, and biofilm formation in Rhizobium leguminosarum bv. viciae 3841. FEMS Microbiol Ecol 71:327–40
- Vigonsky E, Ovcharenko E, Lewinson O. (2013). Two molybdate/tungstate ABC transporters that interact very differently with their substrate binding proteins. Proc Natl Acad Sci USA 110:5440–5
- Voss NR, Gerstein M. (2010). 3V: cavity, channel and cleft volume calculator and extractor. Nucleic Acids Res 38:W555–62
- Walker JE, Saraste M, Runswick MJ, Gay NJ. (1982). Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1:945–51
- Wang T, Fu G, Pan X, et al. (2013). Structure of a bacterial energy-coupling factor transporter. Nature 497:272–6
- Woo JS, Zeltina A, Goetz BA, Locher KP. (2012). X-ray structure of the Yersinia pestis heme transporter HmuUV. Nat Struct Mol Biol 19:1310–15
- Xu K, Zhang M, Zhao Q, et al. (2013). Crystal structure of a folate energy-coupling factor transporter from Lactobacillus brevis. Nature 497:268–71
- Yaffe E, Fishelovitch D, Wolfson HJ, et al. (2008). MolAxis: efficient and accurate identification of channels in macromolecules. Proteins 73:72–86
- Yu Y, Zhou M, Kirsch F, et al. (2014). Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters. Cell Res 24:267–77
- Zhang P. (2013). Structure and mechanism of energy-coupling factor transporters. Trends Microbiol 21:652–9
- Zhang P, Wang J, Shi Y. (2010). Structure and mechanism of the S component of a bacterial ECF transporter. Nature 468:717–20

