Abstract
Context: Statistical and clinical reports indicate that betel nut chewing is strongly associated with progression of oral cancer because some ingredients in betel nuts are potential cancer promoters, especially arecoline. Early diagnosis for cancer biomarkers is the best strategy for prevention of cancer progression. Several methods are suggested for investigating cancer biomarkers. Among these methods, gel-based proteomics approach is the most powerful and recommended tool for investigating biomarkers due to its high-throughput. However, this proteomics approach is not suitable for screening biomarkers with molecular weight under 10 KDa because of the characteristics of gel electrophoresis.
Objective: This study investigated biomarkers with molecular weight under 10 KDa in rats with arecoline challenge.
Materials and methods: The centrifuging vials with membrane (10 KDa molecular weight cut-off) played a crucial role in this study. After centrifuging, the filtrate (containing compounds with molecular weight under 10 KDa) was collected and spotted on a sample plate for MALDI-TOF mass spectrometry analysis.
Results: Compared to control, three extra peaks (m/z values were 1553.1611, 1668.2097 and 1740.1832, respectively) were found in sera and two extra peaks were found in heart tissue samples (408.9719 and 524.9961, respectively). These small compounds should play important roles and may be potential biomarker candidates in rats with arecoline.
Discussion and conclusions: This study successfully reports a mass-based method for investigating biomarker candidates with small molecular weight in different types of sample (including serum and tissue). In addition, this reported method is more time-efficient (1 working day) than gel-based proteomics approach (5~7 working days).
Introduction
According to epidemiological and clinical statistics, betel nut chewing is strongly associated with the occurrence of pathological disorders, including heart disease (CitationLin et al., 2008), obesity (CitationLin et al., 2009) and cancer (CitationSoumitra et al., 1991; CitationGupta & Warnakulasuriya, 2002). Several ingredients in betel nuts have been studied in vitro and in vivo for establishing the connection between betel nut chewing and oral cancer. Among these ingredients, arecoline is the most potential initiator for cancer progression. In order to prevent oral cancer induced by betel nut chewing, early diagnosis for the progression of cancer is necessary. Therefore, screening clinical biomarkers relating to the progression of oral cancer plays a critical role in early diagnosis of cancer.
Several methods have been reported in finding cancer biomarkers. Among these methods, proteomics-based approach is the most recommended due to its high-throughput ability (CitationHu & Wong, 2007; CitationMarkridakis & Vladhou, 2010). In general, proteomics-based approach combines several core technologies, including two-dimension (2D) gel electrophoresis, gel staining, enzymatic digestion, mass spectrometry-based analysis and bioinformation. Once identified by proteomics-based approach, the protein(s) will become one of the candidate biomarkers for cancer.
In addition to 2D, several methods have been mentioned for screening biomarker in proteomics approaches, such as isotope-coded affinity tags (ICAT) (CitationGygi et al., 1999) and surface enhance laser desorption/ionization (SELDI) (Wright Jr et al., 1999; CitationHutchens & Yip, 1993). Although these methods are time-efficient, the experimental costs are relatively higher than 2D due to the use of biochips (or proteinchips) during experiments.
However, proteomics-based approach is the most recommended method in investigating biomarkers, it is usually time-consuming. Basically, a proteomics-based approach needs five to several working days to go through. In addition, proteomics-based approach is suitable for investigating protein biomarkers with molecular weight above 10 KDa due to the characteristics of 2D gel technology. Therefore, inability to investigate biomarkers with 10 KDa is one of limitations for gel-based proteomics approach.
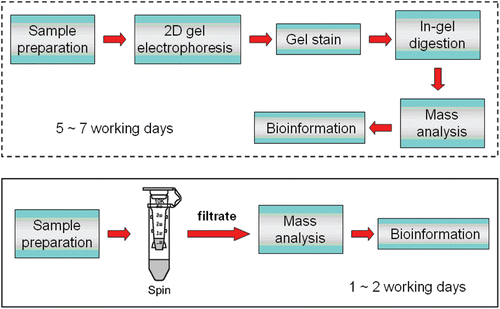
This study focused on avoiding 2D gel electrophoresis, gel staining and enzymatic digestion procedures and screened low molecular weight compounds in arecoline-treated rats by maldi-tof mass spectrometry directly. The use of centrifuging vials with 10 KDa molecular weight cut-off membrane played a critical role in this study. The aim of this study is shown as a flow chart in .
Materials and methods
Chemicals and reagents
All chemicals and reagents used in this study were purchased from Sigma-Aldrich Inc. (St. Louis, MO, USA).
Animal model
Eight week old male SD rats (200~250 g) were divided into two groups including control (n = 8, without arecoline challenge) and treatment (n = 8, with arecoline challenge). The dosage of arecoline for the treatment group was 50 mg/kg and administrated by i.p. injection. All animals were sacrificed after experimental duration of 2 weeks.
Sample preparation
Serum or heart tissue sample (0.2 mL) was transferred into a centrifuging vial with 1000NMWL filter membrane (Amicom Bioseparation, Millipore Corporation, USA). Then all vials were transferred into a centrifuge (Mikro 120, Microlitre Centrifuge, Andreas Hettichi GmbH & Co. KG, Germany), and the centrifuge was set at 1200g.
MALDI-TOF mass spectrometry analysis
After centrifuging, the filtrate was collected for ZipTip C18 treatment (Millipore Corporation, USA). The filtrate after ZipTip treatment was then spotted on a sample plate for maldi-tof MS analysis (Voyager DE STR, Applied Biosystems, USA).
Results
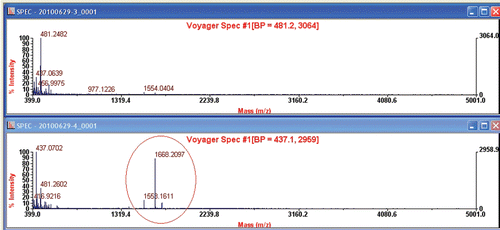
shows spectra for MALDI-TOF MS analysis of sera in rats with or without arecoline treatment. Compared to control (upper spectrum in , mice without arecoline treatment), we can find that three extra peaks were observed in rats with arecoline treatment (lower spectrum in ). The m/z values for these three extra peaks were 1553.1611, 1668.2097 and 1740.1832, respectively.
Figure 2. Spectra for MALDI-TOF MS analysis of sera in rats without (upper spectrum) and with (lower spectrum) arecoline challenge.
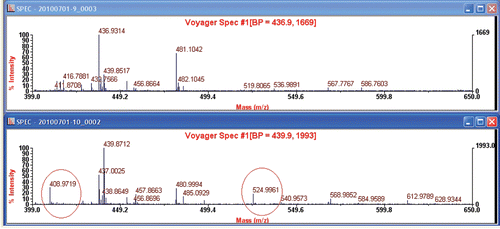
In addition to serum samples, similar results can be observed in tissue samples in rats with or without arecoline treatment. As shown in , compared to control (upper spectrum, heart tissues in rats without arecoline treatment), two extra peaks were presented in the lower spectrum for heart tissues in rats with arecoline treatment. The m/z values for the two extra peaks were 408.9719 and 524.9961, respectively.
Discussion
Proteomics-based approach is a powerful and most recommended tool for screening biomarkers (CitationChambers et al., 2000); however, it is time-consuming (e.g., 2d) or expensive (e.g., ICAT or SELDI). In addition, this approach is suitable for investigating biomarkers with molecular weight above 10 KDa due to the characteristics of 2D gel technology. Therefore, biomarkers with molecular weight under 10 KDa are usually ignored in gel-based proteomics approach. In order to overcome this limitation occurring in gel-based proteomics approach, we used centrifuging vials with molecular weight 10 KDa cut-off membranes to prepare samples. After centrifuging, samples were divided into two parts due to the 10 KDa cut-off membrane, including yogurt-like compounds (with molecular weight above 10 KDa) and filtrate (containing compounds with molecular weight under 10 KDa). The filtrate was further prepared and then was spotted on a sample plate for maldi-tof MS analysis directly. Compared to control, we found that three extra peaks were presented in mass spectrum in rat serum after MALDI-TOF MS analysis (). In addition to serum samples, this mass-based rapid method can be applied in analyzing tissue samples (). These results illustrate that the mass-based method reported in this study is suitable for screening biomarkers with molecular weight under 10 KDa for different samples (including serum and tissue samples). In addition, the experimental procedure for this mass-based method is more time-efficient than gel-based proteomics approach due to avoiding the 2D gel section.
In addition to gel-based proteomics approach, we suggest that this mass-based method may be an alternative method for screening biomarkers with molecular weight under 10 KDa. However, further identification of these biomarker candidates is necessary in future work. For example, de novo protein sequence by tandem mass spectrometry (MSn) or TOF/TOF with bioinformatics should be an appropriate methodology to explore unknown molecules or proteins.
Declaration of interest
This study is supported by CMU98-P-01, CMU98-P-01-M and in part by Taiwan Department of Health Clinical Trial and Research Center of Excellence (DOH101-TD-B-111-004).
References
- Chambers G, Lawrie L, Cash P, Murray GI. (2000). Proteomics: A new approach to the study of disease. J Pathol, 192, 280–288.
- Gupta PC, Warnakulasuriya S. (2002). Global epidemiology of areca nut usage. Addict Biol, 7, 77–83.
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. (1999). Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol, 17, 994–999.
- Hu S, Wong DT. (2007). Oral cancer proteomics. Curr Opin Mol Ther, 9, 467–476.
- Hutchens TW, Yip TT. (1993). New desorption strategies for the mass spectrometric analysis of macromolecules. Rapid Commun Mass Spectrom, 7, 576–580.
- Lin WY, Chiu TY, Lee LT, Lin CC, Huang CY, Huang KC. (2008). Betel nut chewing is associated with increased risk of cardiovascular disease and all-cause mortality in Taiwanese men. Am J Clin Nutr, 87, 1204–1211.
- Lin WY, Pi-Sunyer FX, Liu CS, Li TC, Li CI, Huang CY, Lin CC. (2009). Betel nut chewing is strongly associated with general and central obesity in Chinese male middle-aged adults. Obesity (Silver Spring), 17, 1247–1254.
- Markridakis M, Vladhou A. (2010). Secretome proteomics for discovery of cancer biomarkers. J Proteomics, 73, 2291–305.
- Soumitra S, Geeta T, Archana S. (1991). Betel cytotoxicity: Further evidence from mouse bone marrow cells. Pharm Biol, 29.
- Wright Jr GL, Cazares LH, Leung SM, Nasim S, Adam BL, Yip TT, Schellhammer PF, Gong L, Vlahou A. (1999). Proteinchip® surface enhanced laser desorption/ionization (SELDI) mass spectrometry: A novel protein biochip technology for detection of prostate cancer biomarkers in complex protein mixtures. Prostate Cancer Prostatic Dis, 2, 264–276.