Abstract
According to the structural characteristics of isoliquiritigenin from Glycyrrhiza uralensis, a series of hydroxychalcones has been designed, synthesized and evaluated for their in vitro inhibitory activities of β-secretase (BACE1). Structure-activity relationship study suggested that inhibitory activity against BACE1 was governed to a greater extent by the hydroxyl substituent on A- and B-ring of the chalcone, and the most active compound was substituted with four hydroxyl group (17, IC50 = 0.27 μM).
Introduction
Alzheimer’s disease (AD) is a devastating neurodegenerative disorder characterized by loss of memory and cognition. The accumulation of the β-amyloid peptide (Aβ) in the brain has been thought to be a key factor in the pathogenesis of the diseaseCitation1. Aβ peptides are derived from a sequential proteolytic cleavage of amyloid-precursor protein (APP) by β-secretase (BACE1) and g-secretaseCitation2. The limiting step in this process is cleavage of membrane-bound APP by BACE1Citation1 to form soluble APPCitation3–6. So BACE1 is becoming a molecular target for therapeutic intervention in ADCitation4,Citation7–10. And then numerous BACE1 inhibitors have been synthesized and tested, but most of reported inhibitors have focused on peptide-derived structures, which act as transition state analogs based on the amino acid sequences at the cleavage site of APP by BACE1Citation11. These species showed good activity in italics, but their viability as drug candidates in this case is minimal because the enzyme inhibitors with therapeutic potential are preferably smaller than 700 Da, so large peptide-based inhibitors are not viable drug candidates. Thus, we tried to find the plant-derived non-peptidic inhibitors against BACE1 which would be more likely to be able to reach the target.
With the goal of obtaining new natural inhibitors, we decided to investigate BACE1 inhibitors from the roots of Glycyrrhiza uralensis F., a traditional medicinal herb and an important perennial legume. This herb has long been valued as a demulcent, to relieve respiratory ailments, stomach burn including heart burn, gastritis, inflammatory disorders, skin diseases, and liver problems. Recently, it was reported that G. uralensis water extract markedly improved the cognitive deficits induced in mice by Aβ25–35 administrationCitation12. To look for the biologically active compounds conferring this activity, extract compounds were purified by conventional chromatography and the individual components were assess for BACE1 activity using a BACE1 enzyme-based bioassay. Fortunately, we found isoliquiritigenin (2′, 4′, 4-trihydroxychalcone, 1) showed moderate BACE1 inhibit activity (IC50 = 33.0 μM), and we also found 1 was structurally similar to the natural products catechinsCitation13 and hispidinCitation14 which have also been reported to have BACE1 activity. In this paper, we describe the development of low molecular weight BACE1 inhibitors containing a chalcone skeleton through enzymatic assays.
Chemistry
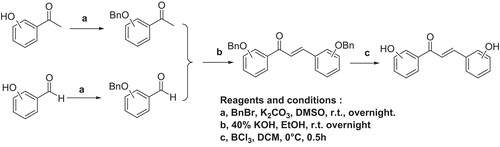
The synthesis of targeted hydroxychalcone analogues (2–22) were achieved using commercially available hydroxyacetophenone and hydroxybenzaldehyde as starting materials, following route as shown in . Hydroxyacetophenone and hydroxybenzaldehyde was benzyl protected under basic conditions, and subsequently reacted in 40% aqueous solution of KOH to form the benzyl protected hydroxychalcone. Then deprotection of the benzyl ether was achieved by treatment with boron trichloride to produce the targeted hydroxychalcone analogues.
Results and discussion
Isoliquiritigenin was prepared to confirm the scaffold compound, and a series of derivatives were synthesized to evaluate their potential in terms of modification of the initial compound, to clarify their structure-activity relationships (SARs).
To determine the best substitution position of a hydroxyl group at B-ring, three compounds with the general structure shown in were designed. The activity results for these compounds are listed in . It seems that BACE1 is sensitive to the position of the substituted group on the B-ring. The result for a hydroxyl substituted at position 2 was superior to those for the hydroxyl substituted at position 3 or position 4. These results indicate the B-ring of isoliquiritigenin (1) for modification to improve the activity of the inhibitor.
Table 1. Importance of the position of a hydroxyl substituent at B-ring.
To further improve the inhibitory potency and determine the effect of the substituted group at position 2, four compounds (3–6) with different substituents at position 2 of B-ring were synthesized. The inhibitory activities of these compounds toward BACE1 are summarized in . To the position 2, the introduction of electron-donating substituents showed better activity than of electron-withdrawing substituents, and the compound 3 with the hydroxyl substituent had the best activity.
Table 2. Diverse compounds at the position 2 of B-ring.
We next turned our attention to modify the A-ring of compound 3. To determine the best substitution position and number of hydroxyl groups at A-ring, nine compounds with the general structure shown in were designed. The activity results for these compounds are listed in . It seems that BACE1 is also sensitive to the substituted positions and numbers of the hydroxyls on the A-ring. The tendency is obvious: the compounds with two hydroxyls (3, 12–16) showed higher inhibitory activity, and the compounds without or with only one hydroxyl (8–11) in ring A show unsatisfactory reduction of the activity of BACE1, so we choose 3, the best activity compound as the beginning compound of the next structure modification.
Table 3. Importance of the positions of hydroxyl substituents at A-ring.
To investigate the SAR of B-ring, six 2-hydroxyl-substituted compounds () were designed. As can be seen from and , the compounds with two hydroxyls (17–22) at B-ring showed better inhibitory activity than those with only one hydroxyl (1–3). Compound 17 with two hydroxyls substituted at the position 2 and 3 showed the best inhibitory activity (IC50 = 0.27 μM).
Table 4. Importance of the positions of hydroxyl substituents at B-ring.
In summary, according to the SARs of all the 22 hydroxychalcones, we found that inhibitory activity against BACE1 was governed to a greater extent by the hydroxyl substituent on A- and B-ring of the chalcone, and the hydroxyl at the position 2, 2′ and 4′ of the chalcone maybe play a critical role in the activity.
Experimental section
General experimental procedures
Unless otherwise mentioned, all chemicals and materials were used as received from commercial suppliers without further purification. Dichloromethane was distilled from calcium hydride. Thin-layer chromatography (TLC) plates (silica gel 60 GF, with glass support) from Yantai jiangyou company were used for monitoring progress of a reaction and visualized with 254 nm UV light. Nuclear magnetic resonance (NMR) spectra were recorded on a Bruker AM-400 spectrometer. EI-MS was obtained on a SHIMADZU GCMS-QP5050A spectrometer.
Extraction and isolation
The air-dried and powdered roots of G. uralensis (2 kg) were percolated with 70% EtOH to afford 130 g of brown extract. Then the extract was suspended in water (500 mL) and further extracted with EtOAc to afford a brown semisolid (33 g after evaporation). The organic layer was subjected to CC (RP-18; MeOH/H2O 60:40) to afford isoliquiritigenin (1) (320 mg).
General procedure for the preparation of compounds (2–22)
To a stirred solution of hydroxyacetophenone (1 mmol) or hydroxybenzaldehyde (0.1 mL, 1 mmol) in dimethyl sulfoxide (DMSO) (10 mL), was added K2CO3 552 mg or 276 mg, respectively. Then, BnBr 0.2 mL (2 mmol) was added to the mixture, and the mixture was stirred at room temperature overnight. After completion of the reaction as indicated by TLC, the mixture was diluted with water and extracted with ethyl acetate (3 × 10 mL). The combined organic layer was washed with brine and dried over Na2SO4. Then, the organic layer was concentrated in a vacuum to afford the corresponding compound of benzyloxyacetophenone or benzyloxybenzaldehyde, which could be used directly for the next step without further purification.
A 40% aqueous solution of potassium hydroxide (5 mL) was added drop-wise to the stirred solution of benzyloxyacetophenone (prepared by the first step) in ethanol (15 mL) under ice-bath. Then, a solution of benzyloxybenzaldehyde in ethanol (10 mL) was added drop-wise to the above mixture, and continued to stir for 30 min under ice-bath, then stirred at room temperature overnight. After completion of the reaction as indicated by TLC, the mixture was concentrated in a vacuum and the residue was diluted with water and extracted with ethyl acetate (3 × 15 mL). The combined organic layer was washed with brine, dried over Na2SO4, and concentrated in a vacuum, the residue was chromatographed on silica gel to afford benzyloxy-chalcone.
To a stirred solution of benzyloxy-chalcone in anhydrous dichloromethane (15 mL) was added drop-wise with 5 mL of BCl3 (1 M in dichloromethane) under ice-bath and the mixture was stirred for 30 min under the same condition. Then ice water (15 mL) was added to the mixture and stirred at room temperature for another 2.5 h. After completion of the reaction as indicated by TLC, the mixture was concentrated in a vacuum and the residue was extracted with ethyl acetate (3 × 15 mL). The combined organic layer was washed with brine, dried over Na2SO4, and concentrated in a vacuum, and the residue was chromatographed on silica gel to afford hydroxylchalcone (2–22).
(E)-1-(2,4-dihydroxyphenyl)-3-(3-hydroxyphenyl)-2-propen-1-one (2)
Yield 82%, yellow solid. Proton-NMR (1H NMR) (DMSO-d6, 400 MHz): δ: 13.4 (1 H, br s), 10.8 (1 H, br s), 9.64 (1 H, br s), 8.19 (1 H, d, J = 8.7), 7.89 (1 H, d, J = 15.6), 7.70 (1 H, d, J = 15.6), 7.23–7.34 (3 H, m), 6.87 (1 H, d, J = 7.5), 6.42 (1 H, d, J = 8.7), 6.30 (1 H, d, J = 2.1); HR-EI-MS: 256.0730 (M+, C15H12O4+; calc. 256.0736).
(E)-1-(2,4-dihydroxyphenyl)-3-(2-hydroxyphenyl)-2-propen-1-one (3)
1H NMR (DMSO-d6, 400 MHz): δ: 13.52 (1 H, s, OH-2), 10.72 (1 H, br s, OH-4), 10.31 (1 H, br s, OH-2′), 8.07 (1 H, d, J = 15.6, H-α), 7.74 (1 H, d, J = 15.6, H-β), 7.78 (1 H, d, J = 8.7, H-6′), 7.51 (1 H, dd, J = 8.7, 2.1, H-6), 7.19 (1 H, td, J = 8.7, 2.7, H-5), 6.85 (1 H, td, J = 8.7, 2.4, H-4), 6.83 (1 H, td, J = 8.7, 2.7, H-3), 6.39 (1 H, dd, J = 8.7, 2.4, H-5′), 6.32 (1 H, d, J = 2.4, H-3′); HR-EI-MS: 256.0731 (M+, C15H12O4+; calc. 256.0736).
(E)-1-(2,4-dihydroxyphenyl)-3-(2-methoxyphenyl)-2-propen-1-one (4)
Yield 83%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 13.5 (1 H, br s), 8.19 (1 H, d, J = 15.6, H-α), 7.84 (1 H, d, J = 9.6), 7.69 (1 H, d, J = 15.6, H-β), 7.63 (1 H, d, J = 7.5), 6.40 (1 H, t, J = 7.5), 7.00 (1 H, d, J = 7.8), 6.96 (1 H, d, J = 8.7), 6.41–6.45 (2 H, m), 3.94 (3 H, s); HR-EI-MS: 270.0899 (M+, C16H14O4+; calc. 270.0892).
(E)-1-(2,4-dihydroxyphenyl)-3-(2-allyloxyphenyl)-2-propen-1-one (5)
Yield 23%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 13.5 (1 H, br s), 10.7 (1 H, br s), 9.28 (1 H, br s), 8.24 (1 H, d, J = 15.0, H-α), 8.16 (1 H, d, J = 8.4), 7.87 (1 H, d, J = 15.6, H-β), 7.83 (1 H, d, J = 7.2), 7.17 (1 H, d, J = 7.2), 6.90 (1 H, t, J = 7.8), 6.41 (1 H, d, J = 9.3), 6.29 (1 H, d, J = 2.4), 5.89–6.00 (1 H, m), 5.06 (1 H, dd, J = 3.3, 1.5), 5.02 (1 H, d, J = 1.2), 3.39 (2 H, d, J = 5.7); HR-EI-MS: 296.1046 (M+, C18H16O4+; calc. 296.1049).
(E)-1-(2,4-dihydroxyphenyl)-3-(2-chlorophenyl)-2-propen-1-one (6)
Yield 85%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 13.2 (1 H, br s), 10.8 (1 H, br s), 8.19–8.25 (2 H, m), 8.10 (1 H, d, J = 15.6, H-α), 8.03 (1 H, d, J = 15.6, H-β), 7.56–7.60 (1 H, m), 7.45–7.51 (2 H, m), 6.43 (1 H, dd, J = 8.7, 2.1), 6.31 (1 H, d, J = 2.7); HR-EI-MS: 274.0391 (M+, C15H11ClO3+; calc. 274.0397).
(E)-1-(2,4-dihydroxyphenyl)-3-(2-nitrophenyl)-2-propen-1-one (7)
Yield 77%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 13.1 (1 H, br s), 10.9 (1 H, br s), 8.20 (2 H, t, J = 6.6), 8.10 (1 H, dd, J = 4.8, 1.5), 8.00 (1 H, d, J = 4.2), 7.83 (1 H, td, J = 5.1, 1.2), 7.70 (1 H, td, J = 5.7, 1.5), 6.45 (1 H, d, J = 5.7), 6.34 (1 H, d, J = 1.2); HR-EI-MS: 285.0642 (M+, C15H11NO5+; calc. 285.0637).
(E)-1-phenyl-3-(2-hydroxyphenyl)-2-propen-1-one (8)
Yield 81%, yellow solid. 1H NMR (CDCl3, 400 MHz): δ: 8.16 (1 H, d, J = 15.6, H-α), 8.04 (2 H, d, J = 6.9), 7.70 (1 H, d, J = 15.6, H-β), 7.60 (1 H, d, J = 7.2), 7.58 (1 H, d, J = 6.6), 7.52 (1 H, d, J = 7.5), 7.49 (1 H, d, J = 6.9), 7.28 (1 H, t, J = 7.5), 6.96 (1 H, t, J = 7.5), 6.91 (1 H, d, J = 8.1), 6.56 (1 H, br s, OH-2′); HR-EI-MS: 224.0833 (M+, C15H12O2+; calc. 224.0837).
(E)-1-(2-hydroxyphenyl)-3-(2-hydroxyphenyl)-2-propen-1-one (9)
Yield 73%, yellow solid. 1H NMR (CDCl3, 400 MHz): δ: 8.20 (1 H, d, J = 15.6, H-α), 7.94 (1 H, d, J = 7.8), 7.85 (1 H, d, J = 15.6, H-β), 7.61 (1 H, d, J = 7.5), 7.50 (1 H, t, J = 7.2), 7.30 (1 H, d, J = 7.2), 7.03 (1 H, d, J = 7.2), 6.98 (1 H, d, J = 8.1), 6.93 (1 H, d, J = 8.1), 6.85 (1 H, d, J = 8.4), 5.73 (1 H, br s); HR-EI-MS: 240.0791 (M+, C15H12O3+; calc. 240.0786).
(E)-1-(3-hydroxyphenyl)-3-(2-hydroxyphenyl)-2-propen-1-one (10)
Yield 75%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 10.3 (1 H, br s), 9.79 (1 H, br s), 8.00 (1 H, d, J = 15.6, H-α), 7.83 (1 H, dd, J = 5.2, 1.2), 7.77 (1 H, d, J = 15.6, H-β), 7.25 (1 H, dt, J = 5.2, 0.9), 7.40 (1 H, m), 7.36 (1 H, t, J = 7.8), 7.27 (1 H, td, J = 7.5, 1.5), 7.04 (1 H, ddd, J = 7.9, 2.4, 0.9), 6.93 (1 H, dd, J = 7.9, 0.9), 6.87 (1 H, t, J = 7.2); HR-EI-MS: 240.0790 (M+, C15H12O3+; calc. 240.0786).
(E)-1-(4-hydroxyphenyl)-3-(2-hydroxyphenyl)-2-propen-1-one (11)
Yield 71%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 10.4 (1 H, br s), 10.2 (1 H, br s), 8.01 (2 H, d, J = 8.4), 7.98 (1 H, d, J = 15.6, H-α), 7.84 (1 H, d, J = 5.4), 7.82 (1 H, d, J = 15.6, H-β), 7.25 (1 H, td, J = 7.8, 1.2), 6.84–6.94 (4 H, m); HR-EI-MS: 240.0793 (M+, C15H12O3+; calc. 240.0786).
(E)-1-(2,3-dihydroxyphenyl)-3-(2-hydroxyphenyl)-2-propen-1-one (12)
Yield 61%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 12.7 (1 H, br s), 10.3 (1 H, br s), 9.37 (1 H, br s), 8.16 (1 H, d, J = 15.6, H-α), 7.89 (1 H, d, J = 9.9), 7.93 (1 H, d, J = 15.6, H-β), 7.63 (1 H, d, J = 8.1), 7.30 (1 H, t, J = 6.9), 7.07 (1 H, d, J = 7.8), 6.95 (1 H, d, J = 8.1), 6.89 (1 H, t, J = 7.8), 6.82 (1 H, t, J = 7.8); HR-EI-MS: 256.0733 (M+, C15H12O4+; calc. 256.0736).
(E)-1-(2,5-dihydroxyphenyl)-3-(2-hydroxyphenyl)-2-propen-1-one (13)
Yield 68%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 11.9 (1 H, br s), 10.4 (1 H, br s), 9.20 (1 H, br s), 8.08 (1 H, d, J = 15.6, H-α), 7.82 (1 H, d, J = 8.7), 7.86 (1 H, d, J = 15.6, H-β), 7.42 (1 H, d, J = 2.7), 7.29 (1 H, t, J = 7.8), 7.02 (1 H, dd, J = 9.0, 2.7), 6.82–6.96 (3 H, m); HR-EI-MS: 256.0732 (M+, C15H12O4+; calc. 256.0736).
(E)-1-(2,6-dihydroxyphenyl)-3-(2-hydroxyphenyl)-2-propen-1-one (14)
Yield 63%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 11.2 (2 H, br s), 10.2 (1 H, br s), 7.89 (1 H, d, J = 15.6, H-α), 7.58 (1 H, d, J = 7.8), 7.79 (1 H, d, J = 15.6, H-β), 7.26 (1 H, t, J = 7.2), 7.20 (1 H, t, J = 8.1), 6.92 (1 H, d, J = 8.1), 6.86 (1 H, t, J = 7.8), 6.38 (2 H, d, J = 8.1); HR-EI-MS: 256.0729 (M+, C15H12O4+; calc. 256.0736).
(E)-1-(3,4-dihydroxyphenyl)-3-(2-hydroxyphenyl)-2-propen-1-one (15)
Yield 59%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 10.2 (2 H, br s), 9.91 (1 H, br s), 9.39 (1 H, br s), 7.94 (1 H, d, J = 15.6, H-α), 7.81 (1 H, d, J = 6.3), 7.78 (1 H, d, J = 15.6, H-β), 7.56 (1 H, d, J = 8.4), 7.48 (1 H, br s), 7.25 (1 H, d, J = 7.2), 6.92 (1 H, t, J = 8.1), 6.87 (1 H, d, J = 5.7), 6.85 (1 H, d, J = 8.1); HR-EI-MS: 256.0733 (M+, C15H12O4+; calc. 256.0736).
(E)-1-(3,5-dihydroxyphenyl)-3-(2-hydroxyphenyl)-2-propen-1-one (16)
Yield 65%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 10.2 (1 H, br s), 9.60 (2 H, br s), 7.93 (1 H, d, J = 15.6, H-α), 7.77 (1 H, d, J = 7.8), 7.66 (1 H, d, J = 15.6, H-β), 7.26 (1 H, t, J = 6.9), 6.84–6.98 (4 H, m), 6.47 (1 H, br s); HR-EI-MS: 256.0738 (M+, C15H12O4+; calc. 256.0736).
(E)-1-(2,4-dihydroxyphenyl)-3-(2,3-dihydroxyphenyl)-2-propen-1-one (17)
Yield 54%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 13.5 (1 H, br s), 8.15 (1 H, d, J = 15.6, H-α), 8.09 (1 H, d, J = 9.0), 7.83 (1 H, d, J = 15.6, H-β), 7.35 (1 H, d, J = 7.8), 6.87 (1 H, d, J = 7.8), 6.71 (1 H, t, J = 7.8), 6.41 (1 H, dd, J = 9.0, 1.5), 6.29 (1 H, d, J = 1.2); EI-MS: 272 [M]+.
(E)-1-(2,4-dihydroxyphenyl)-3-(2,4-dihydroxyphenyl)-2-propen-1-one (18)
Yield 51%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 13.8 (1 H, br s), 8.07 (1 H, d, J = 15.6, H-α), 8.05 (1 H, d, J = 9.0), 7.68 (1 H, d, J = 15.6, H-β), 7.73 (1 H, d, J = 8.4), 6.37–6.41 (2 H, m), 6.32 (1 H, dd, J = 7.2, 2.4), 6.26 (1 H, d, J = 2.1); HR-EI-MS: 272.0681 (M+, C15H12O5+; calc. 272.0685).
(E)-1-(2,4-dihydroxyphenyl)-3-(2,5-dihydroxyphenyl)-2-propen-1-one (19)
Yield 47%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 13.5 (1 H, br s), 10.7 (1 H, br s), 9.61 (1 H, br s), 8.91 (1 H, br s), 8.05 (1 H, d, J = 15.6, H-α), 8.09 (1 H, d, J = 8.4), 7.77 (1 H, d, J = 15.6, H-β), 7.22 (1 H, br s), 6.75 (1 H, d, J = 0.9), 6.41 (1 H, dd, J = 9.0, 2.4), 6.28 (1 H, d, J = 2.7); HR-EI-MS: 272.0682 (M+, C15H12O5+; calc. 272.0685).
(E)-1-(2,4-dihydroxyphenyl)-3-(2,6-dihydroxyphenyl)-2-propen-1-one (20)
Yield 49%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 8.45 (1 H, d, J = 15.6, H-α), 7.83 (1 H, d, J = 9.0), 8.25 (1 H, d, J = 15.6, H-β), 7.03 (1 H, t, J = 8.1), 6.50 (1 H, d, J = 8.1), 6.46 (1 H, dd, J = 9.0, 2.1), 6.36 (1 H, d, J = 2.7); HR-EI-MS: 272.0678 (M+, C15H12O5+; calc. 272.0685).
(E)-1-(2,4-dihydroxyphenyl)-3-(3,4-dihydroxyphenyl)-2-propen-1-one (21)
Yield 49%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 7.73 (1 H, d, J = 8.4), 7.65 (1 H, d, J = 15.3), 7.32 (1 H, d, J = 15.3), 7.08 (1 H, s), 7.00 (1 H, d, J = 8.4), 6.76 (1 H, d, J = 8.4), 6.33 (1 H, dd, J = 8.4, 1.8), 6.28 (1 H, d, J = 1.8); HR-EI-MS: 272.0688 (M+, C15H12O5+; calc. 272.0685).
(E)-1-(2,4-dihydroxyphenyl)-3-(3,5-dihydroxyphenyl)-2-propen-1-one (22)
Yield 55%, yellow solid. 1H NMR (DMSO-d6, 400 MHz): δ: 13.4 (1 H, br s), 10.7 (1 H, br s), 9.49 (2 H, br s), 7.78 (1 H, d, J = 15.6, H-α), 8.17 (1 H, d, J = 9.0), 7.58 (1 H, d, J = 15.6, H-β), 6.70 (2 H, s), 6.28–6.42 (3 H, m); HR-EI-MS: 272.0687 (M+, C15H12O5+; calc. 272.0685).
Biochemical assay
The biochemical assay was employed to determine the IC50 of compounds. The assay based on fluorescence-resonance energy transfer was carried out with BACE1 enzyme at pH 4.5 with a substrate, H-Lys(DABSYL)-SEVNLDAEFR-Gin-(LY) (Merck) according to Ermolieff’s approachCitation15. Briefly, the assay was carried out by incubation of the substrate peptide (5 µM), BACE1 enzyme (550 ng/mL) that was expressed and purified from insect expression system and varied concentrations of compounds at pH 4.5. After 1 h incubation at 37°C, stop buffer (3 M sodium acetate) was introduced into the reaction mixture to stop the reaction. Finally, the fluorescence intensity was measured on a TECAN GENios reader (Tecan) at room temperature with excitation at 420 nm and emission at 530 nm.
Declaration of interest
This work was supported by the Chinese National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program” (grants 2009ZX09301-001, 2009ZX09102-022), the Chinese National High-Tech R&D Program (grants 2007AA02Z147), the National Natural Science Foundation of China (grants 90713046, 30772638, 30925040), CAS Foundation (grant KSCX2-YW-R-179) and China 111 Project (B07023).
References
- Selkoe DJ. Alzheimer’s disease: genes, proteins, and therapy. Physiol Rev 2001;81:741–766.
- Yan R, Bienkowski MJ, Shuck ME, Miao H, Tory MC, Pauley AM, Brashier JR, Stratman NC, Mathews WR, Buhl AE, Carter DB, Tomasselli AG, Parodi LA, Heinrikson RL, Gurney ME. Membrane-anchored aspartyl protease with Alzheimer’s disease beta-secretase activity. Nature 1999;402:533–537.
- Sinha S, Anderson JP, Barbour R, Basi GS, Caccavello R, Davis D, Doan M, Dovey HF, Frigon N, Hong J, Jacobson-Croak K, Jewett N, Keim P, Knops J, Lieberburg I, Power M, Tan H, Tatsuno G, Tung J, Schenk D, Seubert P, Suomensaari SM, Wang S, Walker D, Zhao J, McConlogue L, John V. Purification and cloning of amyloid precursor protein beta-secretase from human brain. Nature 1999;402:537–540.
- Hussain I, Powell D, Howlett DR, Tew DG, Meek TD, Chapman C, Gloger IS, Murphy KE, Southan CD, Ryan DM, Smith TS, Simmons DL, Walsh FS, Dingwall C, Christie G. Identification of a novel aspartic protease (Asp 2) as beta-secretase. Mol Cell Neurosci 1999;14:419–427.
- Lin X, Koelsch G, Wu S, Downs D, Dashti A, Tang J. Human aspartic protease memapsin 2 cleaves the beta-secretase site of beta-amyloid precursor protein. Proc Natl Acad Sci USA 2000;97:1456–1460.
- Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Beta-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science 1999;286:735–741.
- Ghosh AK, Shin D, Downs B, Koelsch G, Lin X, Ermolieff J, Tang J. Design of potent inhibitors for human brain memapsin 2 (beta-secretase). J Am Chem Soc 2000;122:3522–3523.
- Ghosh AK, Bilcer G, Harwood C, Kawahama R, Shin D, Hussain KA, Hong L, Loy JA, Nguyen C, Koelsch G, Ermolieff J, Tang J. Structure-based design: potent inhibitors of human brain memapsin 2 (beta-secretase). J Med Chem 2001;44:2865–2868.
- Tung JS, Davis DL, Anderson JP, Walker DE, Mamo S, Jewett N, Hom RK, Sinha S, Thorsett ED, John VJ. Design of substrate-based inhibitors of human beta-Secretase. Med Chem 2002;45:259–262.
- Tamamura H, Kato T, Otaka A, Fujii N. Synthesis of potent beta-secretase inhibitors containing a hydroxyethylamine dipeptide isostere and their structure-activity relationship studies. Org Biomol Chem 2003;1:2468–2473.
- Asai M, Hattori C, Iwata N, Saido TC, Sasagawa N, Szabó B, Hashimoto Y, Maruyama K, Tanuma S, Kiso Y, Ishiura S. The novel beta-secretase inhibitor KMI-429 reduces amyloid beta peptide production in amyloid precursor protein transgenic and wild-type mice. J Neurochem 2006;96:533–540.
- Ahn J, Um M, Choi W, Kim S, Ha T. Protective effects of Glycyrrhiza uralensis Fisch. on the cognitive deficits caused by beta-amyloid peptide 25–35 in young mice. Biogerontology 2006;7:239–247.
- Jeon SY, Bae K, Seong YH, Song KS. Green tea catechins as a BACE1 (beta-secretase) inhibitor. Bioorg Med Chem Lett 2003;13:3905–3908.
- Park IH, Jeon SY, Lee HJ, Kim SI, Song KS. A beta-secretase (BACE1) inhibitor hispidin from the mycelial cultures of Phellinus linteus. Planta Med 2004;70:143–146.
- Ermolieff J, Loy JA, Koelsch G, Tang J. Proteolytic activation of recombinant pro-memapsin 2 (pro-beta-secretase) studied with new fluorogenic substrates. Biochemistry 2000;39:12450–12456.
- Stachel SJ, Coburn CA, Steele TG, Jones KG, Loutzenhiser EF, Gregro AR, Rajapakse HA, Lai MT, Crouthamel MC, Xu M, Tugusheva K, Lineberger JE, Pietrak BL, Espeseth AS, Shi XP, Chen-Dodson E, Holloway MK, Munshi S, Simon AJ, Kuo L, Vacca JP. Structure-based design of potent and selective cell-permeable inhibitors of human beta-secretase (BACE-1). J Med Chem 2004;47:6447–6450.