Abstract
Acetylpolyamine and spermine oxidases are involved in the catabolism of polyamines. The discovery of selective inhibitors of these enzymes represents an important tool for the development of novel anti-neoplastic drugs. Here, a comparative study on acetylpolyamine and spermine oxidases inhibition by the polyamine analogue chlorhexidine is reported. Chlorhexidine is an antiseptic diamide, commonly used as a bactericidal and bacteriostatic agent. Docking simulations indicate that chlorhexidine binding to these enzymes is compatible with the stereochemical properties of both acetylpolyamine oxidase and spermine oxidase active sites. In fact, chlorhexidine is predicted to establish several polar and hydrophobic interactions with the active site residues of both enzymes, with binding energy values ranging from −7.6 to −10.6 kcal/mol. In agreement with this hypothesis, inhibition studies indicate that chlorhexidine behaves as a strong competitive inhibitor of both enzymes, values of Ki being 0.10 μM and 0.55 μM for acetylpolyamine oxidase and spermine oxidase, respectively.
Introduction
The polyamines (PA) putrescine (Put), spermidine (Spd), and spermine (Spm) are natural aliphatic polycations ubiquitous in living organisms and essential for cell growth, proliferation, and differentiationCitation1,Citation2. In animals, the polyamine catabolism is a recycling pathway that converts Spm to Spd and Spd to Put, with the production of toxic aldehydes and H2O2. Polyamine catabolism is achieved via the concerted action of spermidine/spermine N1-acetyltransferase and acetylpolyamine oxidase (APAO), or alternatively, via spermine oxidase (SMO) with no need of the acetylation stepCitation3–6. In the last decade, APAO and SMO catabolic enzymes have been extensively characterized and well documented to play an essential role in maintaining vertebrate PA homeostasis, the latter being mandatory for cellular lifeCitation1,Citation2,Citation7–10. The recombinant protein expression of the mammalian APAO and SMO in different heterologous systems provided the basis for a deeper understanding of the structure/function relationships of these enzymesCitation11–16. At present, only the three-dimensional structures of two members of the polyamine oxidase family are available, namely the maize polyamine oxidase (ZmPAOCitation17,Citation18) and the yeast polyamine oxidase (FMS1Citation19). Molecular modeling of mouse SMO based on the three-dimensional structure of ZmPAO and sequence analysis of animal polyamine oxidases indicated that the general features of the ZmPAO three-dimensional structure are conserved in the mammalian enzymesCitation11,Citation20,Citation21. Furthermore, molecular modeling, docking and site-directed mutagenesis studies demonstrated that Spm binds to mammalian SMO in the same fashion as observed in the yeast FMS1, which shares with SMO and APAO the substrate oxidation modeCitation21.
Several inhibitors of APAO and SMO have been studied, the most prominent example being MDL 72527 (N1,N4-bis(2,3-butadienyl)-1,4-butanediamine) (), that selectively inhibits FAD-containing polyamine oxidasesCitation14,Citation16,Citation20,Citation22. In particular, APAO and SMO inhibition by 1,8-diaminooctane, 1,12-diaminododecane, N-prenylagmatine and guazatine has been reportedCitation20. Since selective mammalian APAO and SMO inhibitors represent important tools for studying the involvement of this class of enzymes in PA homeostasis and in designing new antineoplastic drugs, the search for new PA analogues with enzyme inhibitory activity results to be an exacting research field. Among potential inhibitor candidates, the present study has focused on the diamide chlorhexidine, N′,N′′′′′-hexane-1,6-diylbis[N-(4 chlorophenyl-)(imidodicarbonimidic diamide)], hereafter referred to as CHX ().
CHX is mainly used in dental applications as an active ingredient in mouthwash, designed to kill dental plaque and other oral bacteria. Actually, CHX based products are usually utilized for the prevention and treatment of periodontal diseases. CHX has been demonstrated to inhibit several host cell-derived proteolytic matrix metallo-proteinases (MMPs), as well as some gingipains (HRgpA, RgpB, and Kgp), which play a major role in periodontitisCitation23,Citation24. However, CHX is also used in non-dental applications, for example in general skin cleaning, as a surgical scrub, and in pre-operative skin preparation.
In this study, we first explored the hypothesis that CHX could act as a polyamine oxidase inhibitor by docking simulations. Docking of CHX in APAO and SMO active sites indicates that CHX binding to these enzymes is compatible with their stereochemical properties. Indeed, CHX is predicted to establish polar and hydrophobic interactions with the active site residues of both enzymes, with binding energy values in the 7.6–10.6 kcal/mol range, which correspond to Kd values ranging from 0.02 µM to 0.6 µM. In agreement with the docking studies, enzyme inhibition analyses indicate that CHX behaves as a strong competitive inhibitor of both enzymes, Ki values being 0.10 µM and 0.55 µM for APAO and SMO, respectively.
Methods
Chemicals
Chlorhexidine (CHX) digluconate solution, spermine (Spm) tetrahydrochloride, N1-acetyl-spermine (N1-acetyl-Spm) trihydrochloride, 4-aminoantipyrine, 3,5-dichloro-2-hydroxybenzenesulfonic acid and horseradish peroxidase were from Sigma-Aldrich (St. Louis, MO, USA). Other compounds were from Sigma-Aldrich (St. Louis, MO, USA), Bio-Rad (Bio-Rad Italia, Milano, Italy) and J.T. Baker (Baker Italia, Milano, Italy). All chemicals were of analytical or reagent grade and used without further purification.
Molecular docking simulations of APAO- and SMO-CHX complexes
Docking of CHX onto the three-dimensional models of APAO and SMO was carried out using AutoDock VinaCitation25. The maximum number of binding modes to generate (maximum number of poses to be returned) was kept to the default value of 9, while the exhaustiveness of the search was increased from default value 8 to 30. The three-dimensional models of APAO and SMO were obtained as described previouslyCitation20,Citation21.
Expression and purification of APAO and SMO proteins in E. coli cells
The recombinant APAO and SMO proteins were expressed in E. coli BL21 DE3 cells and purified as previously reported by Bianchi et al.Citation20 and Cervelli et al.,Citation11 respectively. SDS/PAGE electrophoretic analysis was performed on purified recombinant proteins to assess the grade of purity. Protein concentration was calculated by means of the 460 nm molar extinction coefficient (ϵ460 = 9000 M−1·cm−1) which accounts for FAD absorption.
Enzyme activity and inhibition assays
The APAO and SMO activities were measured spectrophotometrically by following the formation of a pink adduct (ϵ515 nm = 2.6 × 104 M−1 cm−1) as the result of the H2O2-dependent oxidation of 4-aminoantipyrine catalyzed by horseradish peroxidase, and the subsequent condensation of oxidized 4-aminoantipyrine with 3,5-dichloro-2-hydroxybenzenesulfonic acid, as described in Bianchi et al.Citation20 APAO activity was assayed at pH 7.5 (0.1 M Tris-HCl buffer) and 25°C with N1-acetyl-Spm as the substrate in the presence and absence of CHX. SMO activity was assayed at pH 8.3 (0.1 M Tris-HCl buffer) and 25°C using Spm as the substrate in the presence and absence of CHX. Data reported are the average of three different experiments, each with two replicates, standard deviation (SD) was 5%. Under all the experimental conditions, the initial reaction rate for the APAO- and SMO-catalyzed conversion of the substrate was unaffected by the enzyme/inhibitor (i.e. APAO/ and SMO/CHX) incubation time, prior to substrate addition.
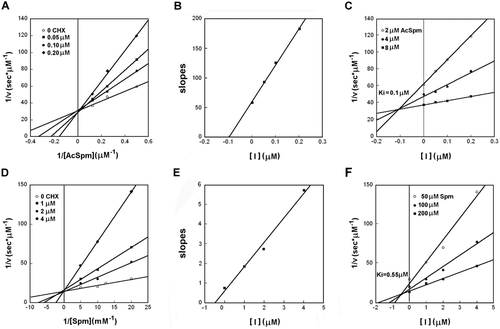
To diagnose the inhibition type, both Lineweaver–Burk (1/v versus 1/s) and Dixon (1/v versus [I]) plots were scrutinized. Estimates of Ki were obtained by second plot of slopes from Lineweaver–Burk versus CHX concentrations and from Dixon plots. In the latter case, two sets of equations were sequentially solved; one equation for each of the straight lines, as obtained by linear least-squares regression, was equated with the line that represented the highest substrate concentration, that line having the smallest slope. The point of intersection of these pairs of lines represents the value of Ki, reported as means.
Results and discussion
Structural bases for inhibition of APAO and SMO by CHX
To probe the hypothesis that CHX could act as a strong competitive inhibitor of APAO and SMO, the molecule was docked inside the two enzymes active sites using the previously published three-dimensional models of SMO and APAOCitation20,Citation21 and the AutoDock Vina molecular docking softwareCitation25. Analysis of the best docking poses highlights a general stereochemical compatibility of CHX with the active sites of both enzymes. Indeed, CHX displays the proper size to fit in the tunnel-shaped active site of animal polyamine oxidases, occupying their entire length (). Several polar and hydrophobic interactions can be observed between CHX and active site residues. In detail, in APAO, one end of the CHX molecule binds in the innermost part of the catalytic tunnel, in the vicinity of the FAD cofactor, thus establishing hydrophobic interactions with Trp62 (). Moving towards the solvent accessible side of the active site, CHX is further stabilized by a charge-charge interaction with Glu89, hydrogen bonds with Asn91 and Arg124, and aromatic stacking interactions with Tyr127 (). The CHX binding mode in SMO is very similar to that observed in APAO, as far as the innermost part of the molecule is concerned. However, moving towards the active site entrance, differences between APAO and SMO active site composition lead to a distinct set of interactions. Glu216 forms a charge-charge interaction with one of the carbonimidic groups and Gln199 establishes a hydrogen bond with one of the imide groups of CHX (). Furthermore, at variance with the APAO-CHX putative complex, the second chlorophenyl group of CHX is solvent inaccessible and bound in a hydrophobic pocket made up by Met197 and Tyr147 ().
Figure 2. Schematic representation of the APAO and SMO best docking complexes with CHX (top panels) and MDL 72527 (bottom panels) showing proteins residues interacting with the inhibitors. The FAD cofactor (colored in magenta) and the active site His residue (His64 and His82 in APAO and SMO, respectively) are shown for reference.
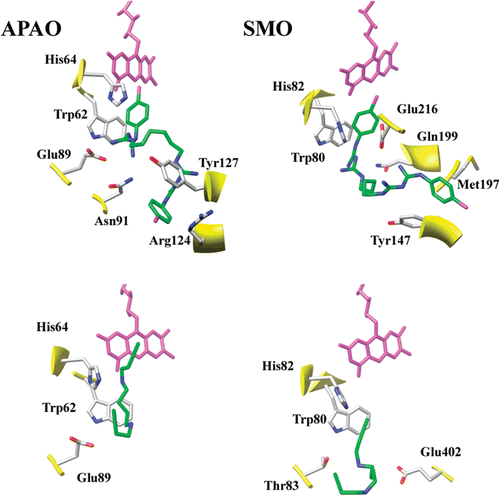
Binding energy values calculated by AutoDock force field for the best nine poses (the default number of poses returned) range from −10.6 to −7.6 kcal/mol. This binding energy values translate in Kd values ranging from 0.02 µM to 0.6 µM, thus suggesting fairly strong inhibition properties of CHX towards animal polyamine oxidases.
To probe for the reliability of AutoDock Vina, docking simulations were performed also with the reference polyamine oxidases inhibitor MDL 72527, known to be a weak, non selective inhibitor of both APAO and SMO [Ki values being 21.0 and 63.0 µM, respectivelyCitation20;]. Analysis of the best docking poses indicates that binding of MDL 72527 to both APAO and SMO takes place essentially only through a hydrophobic interaction with Trp62 side-chain, with the inhibitor binding deeper in the active site in the case of APAO with respect to SMO. In both cases, weak electrostatic interactions are observed between the amino groups of MDL 72527 and polar residues of the enzymes active site. In detail, in APAO, Glu89 is at 4.5 Å distance from one of the MDL 72527 amino group, while in SMO the hydroxyl groups of Thr83 and Glu402 are at 3.6 and 4.5 Å distance from one of the amino groups of MDL 72527, respectively. Binding energy values calculated by AutoDock force field for the best nine poses range from −4.6 to −4.1 kcal/mol, significantly higher than those obtained for CHX, pointing to stronger inhibition properties of the latter compound with respect to MDL 72527.
Inhibition of APAO and SMO by chlorexidine
Purified recombinant murine APAO and SMO follow classical Michaelis–Menten kinetics. Values of Km and kcat for N1-acetyl-spermine (N1-acetyl-Spm) oxidation by APAO are 2.0 µM and 2.0 s−1, respectively. Values of Km and kcat for Spm oxidation by SMO are 90 µM and 4.5 s−1, respectively. The polyamine analogue CHX was tested as in vitro modulator of APAO and SMO activities.
CHX impairs both APAO and SMO activities competitively. Lineweaver–Burk and Dixon plots of APAO and SMO activity inhibition by CHX, as shown in , indicate that CHX behaves as a strong competitive inhibitor of both enzymes. The Ki values were obtained by secondary plotting of Lineweaver–Burk data (slopes versus CHX concentrations) and by Dixon plots, yielding similar results. The obtained values were calculated to be 0.10 ± 0.05 µM and 0.55 ± 0.02 µM for APAO and SMO, respectively ().
Figure 3. Lineweaver–Burk plots reporting reciprocal rate oxidation of N1-acetyl-Spm by APAO (A) and Spm by SMO (D) versus reciprocal substrate concentrations, in the presence and absence of CHX. To calculate Ki for the inhibition by CHX, the data are also reported by second plot of slopes from Lineweaver–Burk plot versus CHX concentrations (B, APAO; E, SMO), and by Dixon plots reporting reciprocal rate substrate oxidation as a function of CHX concentrations (C, APAO; F, SMO)
In agreement with docking simulations results, Ki values for APAO and SMO reported in , clearly demonstrate that CHX is a stronger inhibitor than the reference polyamine oxidases inhibitor MDL 72527. Analysis of the best docking complexes indicates that this is due to the ability of CHX to establish a higher number of interactions with the active site residues of both APAO and SMO. In fact, interaction of MDL 72527 with both enzymes is essentially mediated only by a non-specific hydrophobic interaction with Trp62 (, numbering of APAO) and weak electrostatic interactions with one (in APAO) or two (in SMO) polar residues of the active site. On the contrary, CHX is able to establish a network of interactions both of hydrophobic and electrostatic nature, due to the presence of the chlorophenyl ring and of positively charged carbonimidic and imidic groups. According to docking simulations results, the different binding mode of CHX to APAO with respect to SMO is related to the presence of a hydrophobic cavity at the entrance of the active site in the former enzyme (made up by Tyr147 and Met197) which is absent in the latter. This difference could be exploited to design more effective and more selective inhibitors for APAO and SMO. In so much so that the results of this study suggest that an increase in the polarity of one of the CHX phenyl groups (for instance, substituting one chlorophenyl group with a nitrophenyl one) may very likely improve the inhibition properties of the resulting inhibitor for APAO (strengthening the interaction with Arg124); while a decrease in the polarity of one of the phenyl groups (for instance, substituting one chlorophenyl group with a phenyl one) may result in a stronger inhibitor for SMO.
Table 1. Values of Ki for CHX and MDL 72527 binding to APAO and SMO.
Conclusions
Inhibition studies of related enzymes can provide a deeper insight of the structural basis for substrate and/or inhibitor specificity, and help in designing novel and more selective inhibitors. This is of utmost importance in the case of animal polyamine oxidases, in that the availability of selective inhibitors for APAO and SMO would allow the precise role of these enzymes in polyamine metabolism to be dissected. In this regard, here we show that CHX behaves as the strongest known competitive inhibitor of APAO, the previously known as the best inhibitor being guazatine with a Ki value of 0.45 µM20. In addition, while guazatine displays a Ki value for SMO comparable to that for APAO [0.40 µM20;], CHX is a weaker inhibitor for SMO, Ki value for the latter protein being over five times higher than that for APAO (). Thus, CHX is a promising lead compound to develop highly efficient and selective inhibitors for animal polyamine oxidases. From this viewpoint, docking studies provide some clues on how to modify CHX moiety so as to increase selectivity for APAO or SMO. In fact, it appears that increasing the polarity of one of the CHX chlorophenyl rings may turn out in a better inhibitor for APAO, while a poorer inhibitor for SMO.
Acknowledgements
The authors wish to thank the University of Roma Tre for financial support.
Declaration of interest
The authors report no declarations of interest.
References
- Seiler N. Catabolism of polyamines. Amino Acids 2004;26:217–233.
- Wallace HM, Fraser AV, Hughes A. A perspective of polyamine metabolism. Biochem J 2003;376:1–14.
- Casero RA Jr, Pegg AE. Spermidine/spermine N1-acetyltransferase–the turning point in polyamine metabolism. FASEB J 1993;7:653–661.
- Wang Y, Devereux W, Woster PM, Stewart TM, Hacker A, Casero RA Jr Cloning and characterization of a human polyamine oxidase that is inducible by polyamine analogue exposure. Cancer Res 2001;61:5370–5373.
- Vujcic S, Diegelman P, Bacchi CJ, Kramer DL, Porter CW. Identification and characterization of a novel flavin-containing spermine oxidase of mammalian cell origin. Biochem J 2002;367:665–675.
- Wu T, Yankovskaya V, McIntire WS. Cloning, sequencing, and heterologous expression of the murine peroxisomal flavoprotein, N1-acetylated polyamine oxidase. J Biol Chem 2003;278: 20514–20525.
- Agostinelli E, Arancia G, Vedova LD, Belli F, Marra M, Salvi M et al. The biological functions of polyamine oxidation products by amine oxidases: perspectives of clinical applications. Amino Acids 2004;27:347–358.
- Casero RA Jr, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov 2007;6:373–390.
- Amendola R, Cervelli M, Fratini E, Polticelli F, Sallustio DE, Mariottini P. Spermine metabolism and anticancer therapy. Curr Cancer Drug Targets 2009;9:118–130.
- Casero RA, Pegg AE. Polyamine catabolism and disease. Biochem J 2009;421:323–338.
- Cervelli M, Polticelli F, Federico R, Mariottini P. Heterologous expression and characterization of mouse spermine oxidase. J Biol Chem 2003;278:5271–5276.
- Vujcic S, Liang P, Diegelman P, Kramer DL, Porter CW. Genomic identification and biochemical characterization of the mammalian polyamine oxidase involved in polyamine back-conversion. Biochem J 2003;370:19–28.
- Wang Y, Murray-Stewart T, Devereux W, Hacker A, Frydman B, Woster PM et al. Properties of purified recombinant human polyamine oxidase, PAOh1/SMO. Biochem Biophys Res Commun 2003;304:605–611.
- Bellelli A, Cavallo S, Nicolini L, Cervelli M, Bianchi M, Mariottini P et al. Mouse spermine oxidase: a model of the catalytic cycle and its inhibition by N,N1-bis(2,3-butadienyl)-1,4-butanediamine. Biochem Biophys Res Commun 2004;322:1–8.
- Wang Y, Hacker A, Murray-Stewart T, Frydman B, Valasinas A, Fraser AV et al. Properties of recombinant human N1-acetylpolyamine oxidase (hPAO): potential role in determining drug sensitivity. Cancer Chemother Pharmacol 2005;56:83–90.
- Wu T, Ling KQ, Sayre LM, McIntire WS. Inhibition of murine N1-acetylated polyamine oxidase by an acetylenic amine and the allenic amine, MDL 72527. Biochem Biophys Res Commun 2005;326:483–490.
- Binda C, Coda A, Angelini R, Federico R, Ascenzi P, Mattevi A. A 30-angstrom-long U-shaped catalytic tunnel in the crystal structure of polyamine oxidase. Structure 1999;7:265–276.
- Binda C, Angelini R, Federico R, Ascenzi P, Mattevi A. Structural bases for inhibitor binding and catalysis in polyamine oxidase. Biochemistry 2001;40:2766–2776.
- Huang Q, Liu Q, Hao Q. Crystal structures of Fms1 and its complex with spermine reveal substrate specificity. J Mol Biol 2005;348:951–959.
- Bianchi M, Polticelli F, Ascenzi P, Botta M, Federico R, Mariottini P et al. Inhibition of polyamine and spermine oxidases by polyamine analogues. FEBS J 2006;273:1115–1123.
- Tavladoraki P, Cervelli M, Antonangeli F, Minervini G, Stano P, Federico R et al. Probing mammalian spermine oxidase enzyme-substrate complex through molecular modeling, site-directed mutagenesis and biochemical characterization. Amino Acids 2011;40:1115–1126.
- Bey P, Bolkenius FN, Seiler N, Casara P. N-2,3-Butadienyl-1,4-butanediamine derivatives: potent irreversible inactivators of mammalian polyamine oxidase. J Med Chem 1985;28:1–2.
- Gendron R, Grenier D, Sorsa T, Mayrand D. Inhibition of the activities of matrix metalloproteinases 2, 8, and 9 by chlorhexidine. Clin Diagn Lab Immunol 1999;6:437–439.
- Cronan CA, Potempa J, Travis J, Mayo JA. Inhibition of Porphyromonas gingivalis proteinases (gingipains) by chlorhexidine: synergistic effect of Zn(II). Oral Microbiol Immunol 2006;21:212–217.
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455–461.