Abstract
Carbonic anhydrases (CAs, EC 4.2.1.1) are a family of metalloenzymes that catalyze the reversible interconversion of CO2 and HCO3−. Of the 15 isoforms of human (h) α-CA, 12 are catalytic (hCAs I-IV, VA, VB, VI, VII, IX, XII-XIV). The remaining three acatalytic isoforms (hCAs VIII, X and XI) lack the active site Zn2+ and are referred to as CA-related proteins (CA-RPs); however, their function remains elusive. Overall these isoforms are very similar to each other in structure but they differ in their expression and distribution. The favourable properties of hCA II such as fast kinetics, easy expression and purification, high solubility and intermediate heat resistance have made it an attractive candidate for numerous industrial applications. This review highlights the structural similarity and stability comparison among hCAs.
Classification
Carbonic anhydrases (CAs, EC 4.2.1.1) are a family of metalloenzymes that catalyze the reversible hydration/dehydration of carbon dioxide/bicarbonate ionCitation1,Citation2. CAs are found in both prokaryotes and eukaryotes, and are involved in many physiological processes such as respiration, bone resorption, calcification and photosynthesisCitation3.
CAs have been divided into five distinct classes (α, β, γ, δ and ζ). The α-class is found in mammals, β is found in plants and some prokaryotes, γ is present only in archaeabacteria and two other rare classes (that are similar to class β), δ and ζ are found in diatomsCitation4–9. The three main classes (α, β and γ) of CA are structurally dissimilar and are thought to have evolved independently, possibly as a result of convergent evolution.
Structure and function of human CAs
Structure
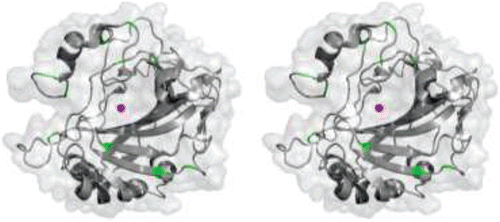
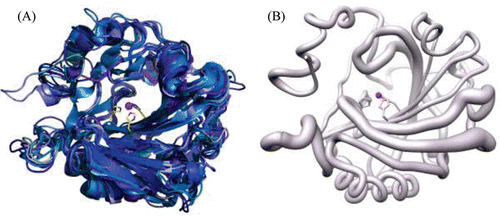
There are 15 isoforms of human (h) α-CA of which, three are acatalytic and will be discussed later. The 12 catalytic isoforms differ from each other in catalytic activity, tissue localization and distribution () in forms of cytosolic (hCAs I-III, VII, XIII), membrane bound (hCAs IV, IX, XII, XIV), secretory (hCA VI) or mitochondrial (hCAs VA, VB)Citation7,Citation10–25. Overall, most hCA isoforms (except hCA VI, IX and XII) exist in monomeric forms composed of seven right handed α-helices, and a twisted β-sheet formed by 10 β-strands (two parallel and eight antiparallel). The CA catalytic domains in transmembrane hCA IX and hCA XII have a similar, but dimeric structure. shows a primary sequence alignment of all the isoforms of hCA, and and summarize the primary sequence identity and structural similarity, respectively. A superposition of all the catalytic isoforms of hCA shows high similarity in the main chain ().
Table 1. Catalytic efficiency and distribution of hCA isoforms.
Table 2. Primary sequence identity (upper right) and number of conserved residues (lower left) among hCA isoforms.
Table 3. Structural main chain r.m.s.d. (Å) among hCA isoforms.
Figure 1. Multiple sequence alignment of the hCA isoforms. Gradients of blue represent sequence conservation from most (dark blue) to least conserved (light blue).

Figure 2. (A) Cartoon representation of superposition of the 11 catalytic isoforms of α-CA, in shades of blue. The histidines (94, 96 and 119) that coordinate the Zn2+ are shown as yellow sticks. (B) Worm representation of main chain r.m.s.d. of the 11 catalytic isoforms. Thicker regions represent more deviation in the main chain. Active site Zn2+ is represented as a magenta sphere. Figure was made using ChimeraCitation110.
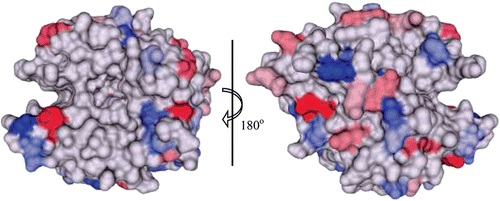
HCA II is the most well studied of all CA isoforms and its active site can be described as a cone-shaped cavity formed of a hydrophobic region (Val121, Val143, Leu198, Val207 and Trp209), and a hydrophilic region (Tyr7, Asn62, His64, Asn67, Thr199 and Thr200). At the base of the cavity lies the Zn2+, tetrahedrally coordinated by three conserved histidines (His94, His96 and His119) and a solvent molecule (). Although the core of the active site in α-CAs is highly conserved, there is variability in the polarity and hydropathicity of its periphery. The conservation of polar and non-polar residues on the surface of catalytic isoforms of hCA is shown in and , respectively.
Figure 3. Stick representation of the active site of hCA II (PDB ID 3D92)Citation2 showing key residues that are involved in CO2 binding and proton transfer during catalysis. Residues are as labelled. Active site Zn2+ is shown as a magenta sphere and water molecules involved in proton transfer are shown as red spheres.
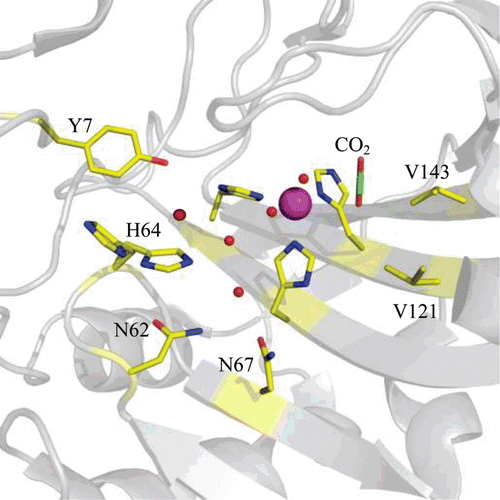
Figure 4. Surface representation of conservation of hydrophobic amino acids among the catalytic α-CAs. The conserved residues are coloured in a gradient of dark (most conserved) to light (least conserved) green. Active site Zn2+ is shown as a magenta sphere. Figure was made using ChimeraCitation110.
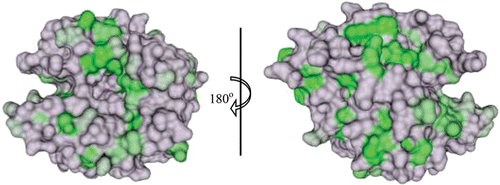
Figure 5. Surface representation of conservation of hydrophilic amino acids among the catalytic α-CAs. The conserved residues are coloured in a gradient of red (negatively charged) and blue (positively charged), from dark (most conserved) to light (least conserved). Active site Zn2+ is shown as a magenta sphere. Figure was made using ChimeraCitation110.
Catalytic mechanism
CAs follow a two-step ping pong catalytic mechanism for the hydration and dehydration of CO2 and HCO3−, respectively:
Step 1: EZnOH− + CO2 ↔ EZnHCO3− ↔ EZnH2O + HCO3−
Step 2: EZnH2O + B ↔ EZnOH− + BH+
The first step of the reaction involves the binding of a CO2 molecule in the active siteCitation2. Molecular dynamics (MD) data suggest the existence of three CO2 binding sites within the active site, one of which is found in the hydrophobic region (3–4 Å away from Zn2+), and the other two are found 6–7 Å away from the Zn2+. Energy analysis suggests that the two binding sites that are 6–7 Å away may act as intermediates guiding CO2 into the hydrophobic regionCitation26. However, the recent X-ray crystal structure (PDB ID: 3D92) reveals a single binding site for CO2 in the active site of hCA II with one of the oxygens of the CO2 interacting with the amide of Thr199 (3.5 Å) and displacing a water molecule whereas, the other oxygen is positioned between the Zn2+ and Val121. This arrangement places both of the oxygens nearly equidistant from the zinc-bound OH− (Zn-OH−), and the carbon 2.8 Å from the Zn-OH−Citation2. Once inside this hydrophobic region, the CO2 undergoes a nucleophillic attack by the Zn-OH−, forming zinc-bound bicarbonate (Zn-HCO3−). This HCO3− is then displaced with a water molecule (Zn-H2O) and the former diffuses into the bulk solvent. In the second step, the Zn-OH− is regenerated as a result of the transfer of a proton from the Zn-H2O generated in step one, to the bulk solvent (or buffer)Citation8.
Kinetic studies show differences in catalytic efficiency among hCAs () which may be attributed to the varying speed of proton shuttling (the rate determining step of catalysis) in step twoCitation26. The proton shuttling step is said to occur by two key events. The first event involves the transfer of the proton via a network of ordered water molecules in the active site to a nearby residue, while the second subsequently releases the proton into the bulk solventCitation27.
One of the key amino acid residues in proton shuttling is His64 (hCA II numbering), which is conserved in all of the catalytic hCAs except III and V. In X-ray crystal structuresCitation28 and MD studiesCitation29, His64 has been found to exist in a dual conformation: inward (7.5 Å away from the Zn2+) and outward (12.0 Å away from the Zn2+) suggesting that the residue acts as a shuttle, accepting the proton then transferring it to the bulk solvent by a flipping mechanism. Although, there exists another hypothesis according to which it is not the flipping of the side chain of His64, but its tautomerism that results in proton transferCitation30. Absence of His64 in hCAs III and V could well be argued as the reason for their relatively slowed catalytic efficiency. However, the rate of proton transfer also depends on the number of water molecules involved in proton transfer, as well as the distance between the proton donor and acceptorCitation31.
In hCA III, mainly three residues are speculated to affect catalysis: Lys64, Arg67 and Phe198 in place of His64, Asn67 and Leu198 (hCA II numbering)Citation32. The loss of His64 for a much more basic Lys, and presence of bulkier Arg67 and Phe198 causes a reduction in the active site cavity volume affecting the binding of CO2, thereby resulting in a lower rate of catalysisCitation33. However, the exact mechanism of how the proton leaves the active site and the exact role of these residues is still unanswered. In CA V, presence of larger residues, Tyr and Phe, in place of His64 and Ala65 (CA II numbering) respectively, makes the region very hydrophobic and thus these residues are believed to be not involved in proton transfer. It is speculated however, that the proton travels through another network possibly involving either Tyr131 or Lys132Citation34.
Up until now, the primary reaction i.e. reversible hydration of carbon dioxide to bicarbonate ion has been discussed. However, CA has been known to be involved in a variety of other reactions (), mechanisms and physiological functions of which are yet to be ascertainedCitation3.
Table 4. Reactions that CAs are believed to be involved in3.
Physiological functions
The isoforms of hCA are found throughout the body in various tissues and sub-cellular locations and have varied physiological roles (). hCAs I, II and III share a high sequence homology, and have been extensively studied. hCAs I, II and IV are involved in the efficient transportation of CO2. hCA II is the most widely expressed isoform and participates in processes ranging from bone resorption to respiration and pH regulation. hCA III is believed to serve an anti-oxidant role in cells which have high rates of oxidation such as adipose tissues, hepatocytes and skeletal muscle fibersCitation35.
hCA V is found in the mitochondrial matrix, existing in two forms: VA and VB. hCA VA provides bicarbonate for gluconeogenesis, and fatty acids for pyrimidine base synthesisCitation3. It is found in the mitochondria of the heart, lung, kidney, spleen and intestines. hCA VB on the other hand, is present in the mitochondria of the pancreas, kidney and salivary glands playing an intermediate role in metabolismCitation36.
hCA VI is the secretory isoform expressed in saliva, milk, nasal secretions and the epithelial lining of digestive organs. The physiological function of hCA VI is yet to be fully understood, but studies show a correlation between the loss of taste and a decrease in hCA VI secretionCitation37. hCA VII (cytosolic) is speculated to play a role in cerebrospinal fluid production in the CNSCitation36. hCA XIII is another cytosolic isoform that is expressed among tissues in the reproductive organs and is speculated to play a role in pH regulation, and ensuring proper fertilizationCitation38.
Among the three transmembrane hCAs, isoforms IX and XII are expressed in the gastrointestinal mucosa. However, they have also been found to be over expressed in epithelial tumours including tumours of the cervix, lungs, kidneys, prostate and breast. In addition, these isoforms are implicated in allowing tumours to acclimate to a hypoxic microenvironment and promoting metastasisCitation39. hCA XIV is present in the brain and retina and is believed to aid in the removal of CO2 from the neural retina and help modulate photoreceptor functionCitation40.
CA-related proteins
The hCA family, besides the catalytic isoforms, includes a subclass of three non-catalytic isoforms (hCAs VIII, X and XI) called CA-related proteins (CA-RPs), based on sequence homology with the catalytic isoforms. The reason behind these isoforms being non-catalytic has been attributed to the absence of one or more histidines that coordinate the Zn2+ ion in the active site of a catalytic hCA isoform. In hCA-RP VIII, for example, the Zn-coordinating His94 (hCA II numbering) is replaced by Arg116 (hCA-RP VIII numbering)Citation41 which precludes CO2 hydration in the first step of CA catalysisCitation42. Although the biological functions of CA-RPs remain undefined, these isoforms continue to gain scientific interest. To date, X-ray crystal structure of only one hCA-RP (hCA-RP VIII) has been determinedCitation43. hCA-RP VIII is highly expressed in the cerebellumCitation44, and has been identified as a binding partner for the inositol 1,4,5 triphosphate (IP3) receptor type 1Citation45.
With a sequence identity of 41%, the overall structure of hCA-RP VIII has an r.m.s.d. of 1.3 Å with the hCA II isoform. The X-ray crystal structure of hCA-RP VIII (PDB ID: 2W2J) reveals a unique glutamate rich loop (E loop; aa 24–36) that is not seen in other hCAs. The core domain of hCA-RP VIII adopts the classical architecture of other α-CAs, namely a 10-stranded central β-sheet flanked by seven short α-helices.
A sequence homology of 98% between murine (m) CA VIII and hCA-RP VIII may well form the hypothesis that over the course of evolution the loss in CA activity is coupled to a gain of a new, though unidentified, cellular function. One possibility is the modulation of biological functions via protein–protein interactions. The type 1 IP3 receptor, identified as a hCA-RP VIII binding protein, contains an electropositive IP3 binding siteCitation46. It is possible that the electronegative surface in hCA-RP VIII, unique among hCAs, forms a charge-complementary binding site for the receptor, thereby regulating IP3-dependent Ca2+ releaseCitation47. Consistent with this, a protein-interaction function has been reported for the CA like domain in receptor protein tyrosine phosphatase β which serves as the contact in binding siteCitation48.
Stability of α-CAs
Stability is an important factor that influences the practical application of a protein. Resistance to various environmental stresses including heat, proteolytic degradation and high concentrations of denaturing agents (e.g. sodium dodecyl sulfate, acid, urea and guanidine-hydrochloride) are some of the factors that define the stability of any protein. Mostly, thermal and chemical stability are strongly correlatedCitation49,Citation50, but this trend is not a valid assumption in many casesCitation51,Citation52. Previous studies involving denaturation and folding of CA have provided an insightful understanding of its folding pathwayCitation8.
Cytosolic hCAs: I and II
Nearly 30 years after the discovery of CA in erythrocytesCitation53, the first detailed denaturation studies on hCAs I and II with concentrated solutions of urea and guanidine-hydrochloride (GuHCl) were conductedCitation54. These and later experiments revealed that isoforms from the same species have varying midpoint concentrations of inactivation (Cm) for GuHCl: 1.5 M for hCA I versus 0.9 M for hCA IICitation55. This also coincides with the same isoform from different species having different stability: bovine (b) CA II denatures in GuHCl at a Cm of 1.6 MCitation56 and has also been shown to be less susceptible to denaturation at high pH than hCA IICitation57,Citation58. With the sequence homology of hCA I, hCA II and bCA II being ~60%, the differences seen in stability between these isoforms seem to arise from as little as two to three amino acid substitutions in the primary sequence. Several studies examining the affect of point mutations in the stability of hCA II in GuHCl show varying destabilization in these variants with Cm values ranging from 0.9 M to <0.1 MCitation59–64.
Early studies involving GuHCl denaturation also revealed that the unfolding pathway of hCA I, hCA II and bCA II occurs in two well-defined transition states via a stable molten-globule intermediate composed of the central β-strandsCitation54,Citation65. Studies involving mutagenesis of residues in this hydrophobic core into radio-labelled cysteines revealed that some portion of this region of hCA II remained folded up to solubility limit of GuHCl (~6 M)Citation59,Citation66,Citation67. Interestingly, the same point mutations mentioned above that showed varying stability of the native structure had little to no effect on the stability of the molten globule state and, in some cases, actually increased the Cm value for the intermediateCitation59–64. Truncation of the first five amino acid residues in the N-terminus of hCA II results in a moderate destabilization (ΔCm = 0.5 M compared to that of wild-type hCA II) of the native stateCitation64. Further truncation of the N-terminus up to 24 residues does not significantly destabilize the native or the molten globule state of the enzyme. This suggests that there are only one or more residues within the first 24 amino acids that interact(s) with the rest of the protein and contribute(s) minimum stabilization energy (~5 kcal/mol)Citation64. In contrast, digestion of the first three C-terminus residues of hCA I with carboxypeptidase revealed a significant decrease in stability from a Cm of 1.5 M GuHCl for the native enzyme to 0.6 M for the truncated variantCitation68.
The metal in the active site of CAs also affects the stability of the enzyme. According to the differential scanning calorimetry (DSC) studies performed on the native and apo-form of hCA II, the enzyme is destabilized by approximately 7°C when comparing the melting temperature (Tm) of the two forms (58°C versus 51°C for native and apo-hCA II, respectively), resulting in a destabilization of about 12 kcal/molCitation69. Cobalt can replace the zinc in the active site of hCA II without major structural changes or significant loss of activity and stabilityCitation70,Citation71 whereas, the Cm-value decreases by approximately 0.2 M GuHCl for cobalt-substituted hCA I and bCA IICitation72 as compared to hCA II.
Studies involving tryptophan fluorescence confirmed the absence of a molten-globule intermediate in the denaturation pathway of hCA II with a Cm value of 4.4 M ureaCitation73. The authors predict that the difference in denaturation between urea and GuHCl is due to the ionic character of GuHCl. The high ionic strength of NaCl may help to weaken the ionic interactions in the native state of hCA II which could explain the similar results seen between denaturation using GuHCl and urea with high salt concentrations.
Circular dichroism (CD) studies performed on hCA I and II suggest approximately 20% α-helix characterization below pH 4, which is much higher than that of the protein near neutral pHCitation74. During the second transition, measurements of the intrinsic viscosity (η) of a solution of bCA II suggests that only partial denaturation occurs at pH 2 (η = 8.4 cm3/g)Citation75, unlike that seen with GuHCl or urea (η = 29.6 cm3/g) which is consistent with the viscosity of a solution containing random coil characteristics (η = 29 cm3/g)Citation76. This partial denaturation in acidic conditions was also observed in hCA I and II with the stability showing a strong dependence on the ionic strength of the bufferCitation58,Citation77. Incomplete denaturation of CA in acidic conditions could be due to electrostatic repulsions that fail to overcome hydrophobic forces, salt bridges and other favourable interactions.
Other cytosolic hCAs: III, VII and XIII
The stability of the other hCAs has not been as extensively studied as hCAs I and II, but what has been reported to date in the literature reveals some similarities and interesting differences between the isoforms. CD studies on bCA III showed a similar profile to that of hCA II, with a molten-globule intermediate occurring at a Cm value of 1.0 M GuHCl and a completely unfolded state at a Cm of 2.6 M GuHClCitation78. Although bCA III contains five free cysteine residues that could form inter- and/or intra-molecular disulphide bridges during the unfolding process, the CD spectra of bCA III in reducing conditions were similar to those without dithiothreitol (DTT). This suggests that these cysteines do not form linkages during denaturation. Interestingly, hydratase and esterase activity studies on CA III isolated from mice (mCA III) at various ages revealed that the mCA III from older mice shows a significant tolerance to thermal denaturation compared to the young miceCitation79. This is believed to be a result of glutathiolation resulting in an unusual disulphide linkage between cysteine residues and glutathione and could provide protection against the highly oxidative environmentCitation79.
The X-ray crystal structure of hCA VII shows a disulphide bond between residues 54 and 178 (hCA VII numbering)Citation80. However, the authors were careful to note that these two cysteines are not conserved in other α-CAs and that disulphide bonds are rare among cytosolic proteins. It was concluded, therefore, that this disulphide bond could be a result of the oxidizing conditions that arise during protein handling. As of present, there have been no stability studies done with hCA VII but, based on the sequence identity to hCA I (51%), hCA II (56%) and hCA III (53%) () similar stability characteristics could be assumed.
Recently, the heat dependency of hCA XIII denaturation has been reportedCitation81. It was shown by DSC studies that hCA XIII undergoes a two-state denaturation phase change from native to a completely unfolded state, similar to those previously reported for hCA ICitation82 and hCA IICitation69,Citation83, with a Tm at approximately 59°C. It was also seen that when complexed with a tight-binding inhibitor of CA, ethoxyzolamide, the Tm increased to 72°C, a common trend seen with other inhibitors of hCA IICitation8.
The extracellular hCAs: IV, VI, IX, XII and XIV
The X-ray crystal structure of hCA IVCitation84 revealed that the presence of two disulphide linkages between 6-11G and 23-203 (hCA IV numbering) may contribute to its stability in 5% SDSCitation85,Citation86. The latter disulphide linkage stabilizes an important loop in the active site containing Thr199, which hydrogen bonds to and orients the zinc-bound hydroxide for catalysisCitation84. A disulphide bridge between residues 23 and 203 was engineered in hCA II. Additionally, Cys206 was mutated to Ser in order to avoid 23–206 disulphide formation. An enhancement in stability of this triple mutant was observed: ▵Cm = 0.8 M GuHClCitation87 and ▵Tm = 13°C (unpublished).
This disulphide (23–203) linkage is conserved in hCAs VI, IX, XII and XIVCitation37,Citation88–90, but these isoforms are more sensitive to SDS than hCA IV. This suggests that the additional disulphide bridge between residues 6 and 11G in hCA IV accounts for the enhanced SDS-resistanceCitation90. Additionally, the X-ray crystal structure of hCA IX reveals a dimeric complex that is linked together via an intermolecular disulphide bridge at residue 41 (hCA IX numbering)Citation88. The implications on stability of this linkage are not yet fully understood, but hCA IX has been associated with tumour cells which are surrounded by a highly acidic environment compared to those of other hCAsCitation91.
The CA-RPs: hCAs VIII, X and XI
Unfolding studies on hCA-RP VIII with GuHCl showed that this isoform, like hCA II, unfolds in two distinct transitions, but was more sensitive to GuHCl chemical denaturation than hCA II (Cm = 0.4 M for hCA-RP VIII, and Cm = 0.9 M for hCA II)Citation41. Based on near-UV CD experiments, it was concluded that the destabilization of hCA-RP VIII could be due to an extended N-terminus, giving it a less compact tertiary structure as compared to hCA II. The sensitivity of hCA-RP VIII to GuHCl could be due to the high ionic character found on the surface (E-loop and α3-β15 loop), as the proposed mechanism of GuHCl denaturation occurs via disruption of ionic charges on the surface of hCA IICitation73. Additionally, the lack of a metal coordinating to the active site has shown to have destabilizing effects in both hCA II and bCA IICitation69,Citation92. Stability studies and high-resolution structures of hCA-RPs X and XI are not yet available, but it may be predicted that the lack of a metal-coordinating ion in the active site of these proteins would have similar destabilizing effects as those seen with hCA-RP VIII, apo-hCA II and bCA II.
Discussion
Recently, the determination of crystal structures for all the catalytically active α-CAs (i.e. hCA ICitation93, hCA IICitation28, hCA IIICitation33, hCA IVCitation84, mCA VACitation34, hCA VI (unpublished, PDB ID 3FE4), hCA VII (unpublished, PDB ID 3MDZ), hCA IXCitation88, hCA XIICitation89, hCA XIIICitation38 and mCA XIVCitation90 will provide a better understanding of the mechanisms of inhibition of different hCAs. This has initiated significant advances in the development of CAIs and their administration in humansCitation94.
Most tumours experience a structurally and functionally disturbed microcirculation of oxygen which pathophysiologically causes an inadequate supply of oxygen (a condition called hypoxia)Citation5,Citation95–100. There is evidence that hCA IX expression allows tumours to acclimate to a hypoxic microenvironment, promoting tumour cell proliferation, and that hCA IX expression is related to poor survival in patientsCitation39,Citation101. hCA IX is a transmembrane protein with a extracellular hCA catalytic domain. This domain is very similar to hCA II and acts as an off-target for most CA inhibitors (CAIs). The only known structure of hCA IX (PDB ID: 3IAI)Citation88 has an r.m.s.d. of 1.5 Å with hCA II (PDB ID: 3KS3)Citation28. However, the existence of certain amino acid differences on the surface, referred to as the “selective pocket”, in hCA IXCitation102 could be exploited for design of isoform specific CAIs. Apart from the catalytic CAs, determination of crystal structure of hCA-RP VIII (PDB ID: 2W2J)Citation43 has reinforced the efforts to study the other two hCA-RPs and investigate the possible role of these acatalytic CA isoforms.
The biophysical properties of CAs that contribute to the stability and folding pathway have been extensively studied. While these studies have mainly focused on hCAs I and II, the conclusions can be applied to other protein families in which these characteristics remain elusive. Further research is needed to fully characterize the stability of several human isoforms, namely the extracellular hCAs (IV, VI, IX, XII and XIV) as well as the mitochondrial hCAs (VA and VB). All of these isoforms experience unique environments compared to those of the cytosolic hCAs (I, II, III, VII and XIII).
The favourable properties of hCA II (high kinetic parameters, easy expression, high solubility, intermediate heat resistance) have made it an attractive candidate for numerous industrial applicationsCitation83; however, there are few prokaryotic extremophilic CAs (such as SspCA and SazCA) recently discovered in Sulfurihydrogenibium yellowstonense that show comparable enzymatic activity and higher thermostabilityCitation103,Citation104. There is an increasing industrial interest in using hCA II as a bio-catalyst for carbon sequestration of flue-gas from coal-fired power plants. Also, there are established protocols utilizing CA found in algae to capture carbon dioxide and convert it into biofuels and other valuable productsCitation105,Citation106. There is also interest in using apo-CAs as a bio-sensor for zinc and other transition metals in sea water or human serumCitation107.
For industrial applications, small improvements in stability without detriment to yield, activity or solubility, can accelerate the development of hCA II as a better bio-catalyst. Use of the free enzyme in solution can also have disadvantages, as the low-stability can limit recycling and cost-efficiency in an industrial settingCitation108. As such, there are numerous studies underway to enhance the stability of hCA II while also retaining its characteristic high catalytic efficiency. One such study deals with mutating some residues to Cys in wild-type hCA II in order to form disulphide linkagesCitation87. shows regions in hCA II where Cys residues can possibly be engineered, based on their presence in other hCAs. Other studies include immobilization of hCA II on a variety of surfacesCitation108,Citation109 and directed-evolution of the enzyme involving mutagenesis of surface hydrophobic residues into hydrophilic moietiesCitation83. Further research is needed to maximize the stability of hCA II in a wide array of environments without the loss of catalytic efficiency for industrial use.
Declaration of interest
The work has been supported by Alumni Fellowship Award, Grinter Award and Medical Guild Research Incentive Award from University of Florida (MA), HHMI Science for Life (BK) and National Institutes of Health grant GM25154 (RM). The authors report no conflict of interest.
References
- Messerchmidt A, Bode W, Cygler M. (2004). Handbook of Metalloproteins. West Sussex, England: John Wiley and Sons Ltd.
- Domsic JF, Avvaru BS, Kim CU, Gruner SM, Agbandje-McKenna M, Silverman DN et al. Entrapment of carbon dioxide in the active site of carbonic anhydrase II. J Biol Chem 2008;283:30766–30771.
- Supuran CT. Carbonic anhydrases–an overview. Curr Pharm Des 2008;14:603–614.
- Scozzafava A, Mastrolorenzo A, Supuran CT. Carbonic anhydrase inhibitors and activators and their use in therapy. Expert Opinion on Therapeutic Patents 2006;16:1627–1664.
- Supuran CT, Scozzafava A, Conway J. (2004). Carbonic anhydrase: its inhibitors and activators. Boca Raton, Florida (USA): CRC Press.
- Pastorekova S, Parkkila S, Pastorek J, Supuran CT. Carbonic anhydrases: current state of the art, therapeutic applications and future prospects. J Enzyme Inhib Med Chem 2004;19:199–229.
- Chegwidden WR, Carter ND. (2000). The carbonic anhydrases: new horizons. Birkhauser Verlag. Basel, Switzerland: Birkhäuser.
- Krishnamurthy VM, Kaufman GK, Urbach AR, Gitlin I, Gudiksen KL, Weibel DB et al. Carbonic anhydrase as a model for biophysical and physical-organic studies of proteins and protein-ligand binding. Chem Rev 2008;108:946–1051.
- Supuran CT, Scozzafava A, Casini A. Carbonic anhydrase inhibitors. Med Res Rev 2003;23:146–189.
- Nishimori I, Minakuchi T, Onishi S, Vullo D, Cecchi A, Scozzafava A et al. Carbonic anhydrase inhibitors: cloning, characterization, and inhibition studies of the cytosolic isozyme III with sulfonamides. Bioorg Med Chem 2007;15:7229–7236.
- Vullo D, Franchi M, Gallori E, Antel J, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors. Inhibition of mitochondrial isozyme V with aromatic and heterocyclic sulfonamides. J Med Chem 2004;47:1272–1279.
- Nishimori I, Vullo D, Innocenti A, Scozzafava A, Mastrolorenzo A, Supuran CT. Carbonic anhydrase inhibitors. The mitochondrial isozyme VB as a new target for sulfonamide and sulfamate inhibitors. J Med Chem 2005;48:7860–7866.
- Nishimori I, Minakuchi T, Onishi S, Vullo D, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors. DNA cloning, characterization, and inhibition studies of the human secretory isoform VI, a new target for sulfonamide and sulfamate inhibitors. J Med Chem 2007;50:381–388.
- Vullo D, Voipio J, Innocenti A, Rivera C, Ranki H, Scozzafava A et al. Carbonic anhydrase inhibitors. Inhibition of the human cytosolic isozyme VII with aromatic and heterocyclic sulfonamides. Bioorg Med Chem Lett 2005;15:971–976.
- Vullo D, Franchi M, Gallori E, Pastorek J, Scozzafava A, Pastorekova S et al. Carbonic anhydrase inhibitors: inhibition of the tumor-associated isozyme IX with aromatic and heterocyclic sulfonamides. Bioorg Med Chem Lett 2003;13:1005–1009.
- Vullo D, Innocenti A, Nishimori I, Pastorek J, Scozzafava A, Pastoreková S et al. Carbonic anhydrase inhibitors. Inhibition of the transmembrane isozyme XII with sulfonamides-a new target for the design of antitumor and antiglaucoma drugs? Bioorg Med Chem Lett 2005;15:963–969.
- Nishimori I, Vullo D, Innocenti A, Scozzafava A, Mastrolorenzo A, Supuran CT. Carbonic anhydrase inhibitors: inhibition of the transmembrane isozyme XIV with sulfonamides. Bioorg Med Chem Lett 2005;15:3828–3833.
- Lehtonen J, Shen B, Vihinen M, Casini A, Scozzafava A, Supuran CT et al. Characterization of CA XIII, a novel member of the carbonic anhydrase isozyme family. J Biol Chem 2004;279:2719–2727.
- Supuran CT, Ilies MA, Scozzafava A. Carbonic anhydrase inhibitors — Part 29 1: Interaction of isozymes I, II and IV with benzolamide-like derivatives. Eur J Med Chem 1998;33:739–751.
- Supuran CT, Scozzafava A, Ilies MA, Briganti F. Carbonic anhydrase inhibitors: synthesis of sulfonamides incorporating 2,4,6-trisubstituted-pyridinium-ethylcarboxamido moieties possessing membrane-impermeability and in vivo selectivity for the membrane-bound (CA IV) versus the cytosolic (CA I and CA II) isozymes. J Enzym Inhib 2000;15:381–401.
- Scozzafava A, Briganti F, Ilies MA, Supuran CT. Carbonic anhydrase inhibitors: synthesis of membrane-impermeant low molecular weight sulfonamides possessing in vivo selectivity for the membrane-bound versus cytosolic isozymes. J Med Chem 2000;43:292–300.
- Winum JY, Temperini C, El Cheikh K, Innocenti A, Vullo D, Ciattini S et al. Carbonic anhydrase inhibitors: clash with Ala65 as a means for designing inhibitors with low affinity for the ubiquitous isozyme II, exemplified by the crystal structure of the topiramate sulfamide analogue. J Med Chem 2006;49:7024–7031.
- Saczewski F, Slawinski J, Kornicka A, Brzozowski Z, Pomarnacka E, Innocenti A et al. Carbonic anhydrase inhibitors. Inhibition of the cytosolic human isozymes I and II, and the transmembrane, tumor-associated isozymes IX and XII with substituted aromatic sulfonamides activatable in hypoxic tumors. Bioorg Med Chem Lett 2006;16:4846–4851.
- De Simone G, Vitale RM, Di Fiore A, Pedone C, Scozzafava A, Montero JL et al. Carbonic anhydrase inhibitors: Hypoxia-activatable sulfonamides incorporating disulfide bonds that target the tumor-associated isoform IX. J Med Chem 2006;49:5544–5551.
- Köhler K, Hillebrecht A, Schulze Wischeler J, Innocenti A, Heine A, Supuran CT et al. Saccharin inhibits carbonic anhydrases: possible explanation for its unpleasant metallic aftertaste. Angew Chem Int Ed Engl 2007;46:7697–7699.
- Liang JY, Lipscomb WN. Binding of substrate CO2 to the active site of human carbonic anhydrase II: a molecular dynamics study. Proc Natl Acad Sci USA 1990;87:3675–3679.
- Lindskog S. Structure and mechanism of carbonic anhydrase. Pharmacol Ther 1997;74:1–20.
- Avvaru BS, Kim CU, Sippel KH, Gruner SM, Agbandje-McKenna M, Silverman DN et al. A short, strong hydrogen bond in the active site of human carbonic anhydrase II. Biochemistry 2010;49:249–251.
- Maupin CM, Castillo N, Taraphder S, Tu C, McKenna R, Silverman DN et al. Chemical rescue of enzymes: proton transfer in mutants of human carbonic anhydrase II. J Am Chem Soc 2011;133:6223–6234.
- Shimahara H, Yoshida T, Shibata Y, Shimizu M, Kyogoku Y, Sakiyama F et al. Tautomerism of histidine 64 associated with proton transfer in catalysis of carbonic anhydrase. J Biol Chem 2007;282:9646–9656.
- Fisher Z, Hernandez Prada JA, Tu C, Duda D, Yoshioka C, An H et al. Structural and kinetic characterization of active-site histidine as a proton shuttle in catalysis by human carbonic anhydrase II. Biochemistry 2005;44:1097–1105.
- Elder I, Fisher Z, Laipis PJ, Tu C, McKenna R, Silverman DN. Structural and kinetic analysis of proton shuttle residues in the active site of human carbonic anhydrase III. Proteins 2007;68:337–343.
- Duda DM, Tu C, Fisher SZ, An H, Yoshioka C, Govindasamy L et al. Human carbonic anhydrase III: structural and kinetic study of catalysis and proton transfer. Biochemistry 2005;44:10046–10053.
- Boriack-Sjodin PA, Heck RW, Laipis PJ, Silverman DN, Christianson DW. Structure determination of murine mitochondrial carbonic anhydrase V at 2.45-A resolution: implications for catalytic proton transfer and inhibitor design. Proc Natl Acad Sci USA 1995;92:10949–10953.
- Mitterberger MC, Kim G, Rostek U, Levine RL, Zwerschke W. Carbonic anhydrase III regulates peroxisome proliferator-activated receptor-?2. Exp Cell Res 2012;318:877–886.
- Imtaiyaz Hassan M, Shajee B, Waheed A, Ahmad F, Sly WS. Structure, function and applications of carbonic anhydrase isozymes. Bioorg Med Chem 2012 (In Press).
- Pilka ES, Kochan G, Oppermann U, Yue WW. Crystal structure of the secretory isozyme of mammalian carbonic anhydrases CA VI: implications for biological assembly and inhibitor development. Biochem Biophys Res Commun 2012;419:485–489.
- Di Fiore A, Monti SM, Hilvo M, Parkkila S, Romano V, Scaloni A et al. Crystal structure of human carbonic anhydrase XIII and its complex with the inhibitor acetazolamide. Proteins 2009;74:164–175.
- Cianchi F, Vinci MC, Supuran CT, Peruzzi B, De Giuli P, Fasolis G et al. Selective inhibition of carbonic anhydrase IX decreases cell proliferation and induces ceramide-mediated apoptosis in human cancer cells. J Pharmacol Exp Ther 2010;334:710–719.
- Nagelhus EA, Mathiisen TM, Bateman AC, Haug FM, Ottersen OP, Grubb JH et al. Carbonic anhydrase XIV is enriched in specific membrane domains of retinal pigment epithelium, Muller cells, and astrocytes. Proc Natl Acad Sci USA 2005;102:8030–8035.
- Bergenhem NC, Hallberg M, Wisén S. Molecular characterization of the human carbonic anhydrase-related protein (HCA-RP VIII). Biochim Biophys Acta 1998;1384:294–298.
- Tashian RE, Hewett-Emmett D, Carter N, Bergenhem NC. Carbonic anhydrase (CA)-related proteins (CA-RPs), and transmembrane proteins with CA or CA-RP domains. EXS 2000:105–120.
- Picaud SS, Muniz JR, Kramm A, Pilka ES, Kochan G, Oppermann U et al. Crystal structure of human carbonic anhydrase-related protein VIII reveals the basis for catalytic silencing. Proteins 2009;76:507–511.
- Lakkis MM, Bergenhem NC, O’Shea KS, Tashian RE. Expression of the acatalytic carbonic anhydrase VIII gene, Car8, during mouse embryonic development. Histochem J 1997;29:135–141.
- Hirota J, Ando H, Hamada K, Mikoshiba K. Carbonic anhydrase-related protein is a novel binding protein for inositol 1,4,5-trisphosphate receptor type 1. Biochem J 2003;372:435–441.
- Bosanac I, Alattia JR, Mal TK, Chan J, Talarico S, Tong FK et al. Structure of the inositol 1,4,5-trisphosphate receptor binding core in complex with its ligand. Nature 2002;420:696–700.
- Jiao Y, Yan J, Zhao Y, Donahue LR, Beamer WG, Li X et al. Carbonic anhydrase-related protein VIII deficiency is associated with a distinctive lifelong gait disorder in waddles mice. Genetics 2005;171:1239–1246.
- Peles E, Schlessinger J, Grumet M. Multi-ligand interactions with receptor-like protein tyrosine phosphatase beta: implications for intercellular signaling. Trends Biochem Sci 1998;23:121–124.
- Willuda J, Honegger A, Waibel R, Schubiger PA, Stahel R, Zangemeister-Wittke U et al. High thermal stability is essential for tumor targeting of antibody fragments: engineering of a humanized anti-epithelial glycoprotein-2 (epithelial cell adhesion molecule) single-chain Fv fragment. Cancer Res 1999;59:5758–5767.
- Yao M, Bolen DW. How valid are denaturant-induced unfolding free energy measurements? Level of conformance to common assumptions over an extended range of ribonuclease A stability. Biochemistry 1995;34:3771–3781.
- Samiotakis A, Homouz D, Cheung MS. Multiscale investigation of chemical interference in proteins. J Chem Phys 2010;132:175101.
- Wang Q, Christiansen A, Samiotakis A, Wittung-Stafshede P, Cheung MS. Comparison of chemical and thermal protein denaturation by combination of computational and experimental approaches. II. J Chem Phys 2011;135:175102.
- Stadie WC, O’Brien H. The catalysis of the hydration of carbon dioxide and dehydration of carbonic acid by an enzyme isolated from red blood cells. J Biol Chem 1933;103:521–529.
- Jagannadham MV, Balasubramanian D. The molten globular intermediate form in the folding pathway of human carbonic anhydrase B. FEBS Lett 1985;188:326–330.
- Carlsson U, Henderson LE, Lindskog S. Denaturation and reactivation of human carbonic anhydrases in guanidine hydrochloride and urea. Biochim Biophys Acta 1973;310:376–387.
- Yazgan A, Henkens RW. Role of zinc (II) in the refolding of guanidine hydrochloride denatured bovine carbonic anhydrase. Biochemistry 1972;11:1314–1318.
- Laurent G, Charrel M, Garcon D, Castay M, Marriq C, Derrien Y. [On the proteins accompanying human hemoglobin in its preparations. 3. Identification of proteins Y, X1 and X2 with erythrocyte carbonic anhydrases and unequal stability of these in an alkaline medium]. Bull Soc Chim Biol 1964;46:603–620.
- Riddiford LM, Stellwagen RH, Mehta S, Edsall JT. Hydrogen ion equilibria of human carbonic anhydrases B and C. J Biol Chem 1965;240:3305–3316.
- Svensson M, Jonasson P, Freskgård PO, Jonsson BH, Lindgren M, Mårtensson LG et al. Mapping the folding intermediate of human carbonic anhydrase II. Probing substructure by chemical reactivity and spin and fluorescence labeling of engineered cysteine residues. Biochemistry 1995;34:8606–8620.
- Mårtensson LG, Jonsson BH, Andersson M, Kihlgren A, Bergenhem N, Carlsson U. Role of an evolutionarily invariant serine for the stability of human carbonic anhydrase II. Biochim Biophys Acta 1992;1118:179–186.
- Mårtensson LG, Jonasson P, Freskgård PO, Svensson M, Carlsson U, Jonsson BH. Contribution of individual tryptophan residues to the fluorescence spectrum of native and denatured forms of human carbonic anhydrase II. Biochemistry 1995;34:1011–1021.
- Fransson C, Freskgård PO, Herbertsson H, Johansson A, Jonasson P, Mårtensson LG et al. Cis-trans isomerization is rate-determining in the reactivation of denatured human carbonic anhydrase II as evidenced by proline isomerase. FEBS Lett 1992;296:90–94.
- Tweedy NB, Nair SK, Paterno SA, Fierke CA, Christianson DW. Structure and energetics of a non-proline cis-peptidyl linkage in a proline-202–>alanine carbonic anhydrase II variant. Biochemistry 1993;32:10944–10949.
- Aronsson G, Mårtensson LG, Carlsson U, Jonsson BH. Folding and stability of the N-terminus of human carbonic anhydrase II. Biochemistry 1995;34:2153–2162.
- Henkens RW, Kitchell BB, Lottich SC, Stein PJ, Williams TJ. Detection and characterization using circular dichroism and fluorescence spectroscopy of a stable intermediate conformation formed in the denaturation of bovine carbonic anhydrase with guanidinium chloride. Biochemistry 1982;21:5918–5923.
- Carlsson U, Aasa R, Henderson LE, Jonsson BH, Lindskog S. Paramagnetic and fluorescent probes attached to “buried” sulfhydryl groups in human carbonic anhydrases. Application to inhibitor binding, denaturation and refolding. Eur J Biochem 1975;52:25–36.
- Mårtensson LG, Jonsson BH, Freskgård PO, Kihlgren A, Svensson M, Carlsson U. Characterization of folding intermediates of human carbonic anhydrase II: probing substructure by chemical labeling of SH groups introduced by site-directed mutagenesis. Biochemistry 1993;32:224–231.
- Carlsson U, Henderson LE, Nyman PO, Samuelsson T. Studies on the influence of carboxyl-terminal amino acid residues on the activity and stability of human erythrocyte carbonic anhydrase B. FEBS Lett 1974;48:167–171.
- Avvaru BS, Busby SA, Chalmers MJ, Griffin PR, Venkatakrishnan B, Agbandje-McKenna M et al. Apo-human carbonic anhydrase II revisited: implications of the loss of a metal in protein structure, stability, and solvent network. Biochemistry 2009;48:7365–7372.
- Håkansson K, Wehnert A, Liljas A. X-ray analysis of metal-substituted human carbonic anhydrase II derivatives. Acta Crystallogr D Biol Crystallogr 1994;50:93–100.
- Hunt JA, Ahmed M, Fierke CA. Metal binding specificity in carbonic anhydrase is influenced by conserved hydrophobic core residues. Biochemistry 1999;38:9054–9062.
- Bergenhem N, Carlsson U, Lind G, Astrand IM. Denaturation and reactivation of bovine and human cobalt–carbonic anhydrases in guanidine hydrochloride. Acta Chem Scand, B, Org Chem Biochem 1983;37:244–246.
- Borén K, Grankvist H, Hammarström P, Carlsson U. Reshaping the folding energy landscape by chloride salt: impact on molten-globule formation and aggregation behavior of carbonic anhydrase. FEBS Lett 2004;566:95–99.
- Beychok S, Armstrong JM, Lindblow C, Edsall JT. Optil rotatory dispersion and circular dichroism of human carbonic anhydrases B and C. J Biol Chem 1966;241:5150–5160.
- Wong KP, Hamlin LM. Acid denaturation of bovine carbonic anhydrase B. Biochemistry 1974;13:2678–2683.
- Wong KP, Tanford C. Denaturation of bovine carbonic anhydrase B by guanidine hydrochloride. A process involving separable sequential conformational transitions. J Biol Chem 1973;248:8518–8523.
- Riddiford LM. Acid difference spectra of human carbonic anhydrases. J Biol Chem 1965;240:168–172.
- Borén K, Andersson P, Larsson M, Carlsson U. Characterization of a molten globule state of bovine carbonic anhydrase III: loss of asymmetrical environment of the aromatic residues has a profound effect on both the near- and far-UV CD spectrum. Biochim Biophys Acta 1999;1430:111–118.
- Cabiscol E, Levine RL. Carbonic anhydrase III. Oxidative modification in vivo and loss of phosphatase activity during aging. J Biol Chem 1995;270:14742–14747.
- Di Fiore A, Truppo E, Supuran CT, Alterio V, Dathan N, Bootorabi F et al. Crystal structure of the C183S/C217S mutant of human CA VII in complex with acetazolamide. Bioorg Med Chem Lett 2010;20:5023–5026.
- Baranauskiene L, Matulis D. Intrinsic thermodynamics of ethoxzolamide inhibitor binding to human carbonic anhydrase XIII. BMC Biophys 2012;5:12.
- Matulis D, Kranz JK, Salemme FR, Todd MJ. Thermodynamic stability of carbonic anhydrase: measurements of binding affinity and stoichiometry using ThermoFluor. Biochemistry 2005;44:5258–5266.
- Fisher Z, Boone CD, Biswas SM, Venkatakrishnan B, Aggarwal M, Tu C et al. Kinetic and structural characterization of thermostabilized mutants of human carbonic anhydrase II. Protein Eng Des Sel 2012;25:347–355.
- Stams T, Nair SK, Okuyama T, Waheed A, Sly WS, Christianson DW. Crystal structure of the secretory form of membrane-associated human carbonic anhydrase IV at 2.8-A resolution. Proc Natl Acad Sci USA 1996;93:13589–13594.
- Whitney PL, Briggle TV. Membrane-associated carbonic anhydrase purified from bovine lung. J Biol Chem 1982;257:12056–12059.
- Waheed A, Okuyama T, Heyduk T, Sly WS. Carbonic anhydrase IV: purification of a secretory form of the recombinant human enzyme and identification of the positions and importance of its disulfide bonds. Arch Biochem Biophys 1996;333:432–438.
- Mårtensson LG, Karlsson M, Carlsson U. Dramatic stabilization of the native state of human carbonic anhydrase II by an engineered disulfide bond. Biochemistry 2002;41:15867–15875.
- Alterio V, Hilvo M, Di Fiore A, Supuran CT, Pan P, Parkkila S et al. Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX. Proc Natl Acad Sci USA 2009;106:16233–16238.
- Whittington DA, Waheed A, Ulmasov B, Shah GN, Grubb JH, Sly WS et al. Crystal structure of the dimeric extracellular domain of human carbonic anhydrase XII, a bitopic membrane protein overexpressed in certain cancer tumor cells. Proc Natl Acad Sci USA 2001;98:9545–9550.
- Whittington DA, Grubb JH, Waheed A, Shah GN, Sly WS, Christianson DW. Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: implications for selective inhibition of membrane-associated isozymes. J Biol Chem 2004;279:7223–7228.
- Wykoff CC, Beasley NJ, Watson PH, Turner KJ, Pastorek J, Sibtain A et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res 2000;60:7075–7083.
- Andersson D, Hammarström P, Carlsson U. Cofactor-induced refolding: refolding of molten globule carbonic anhydrase induced by Zn(II) and Co(II). Biochemistry 2001;40:2653–2661.
- Kannan KK, Ramanadham M, Jones TA. Structure, refinement, and function of carbonic anhydrase isozymes: refinement of human carbonic anhydrase I. Ann N Y Acad Sci 1984;429:49–60.
- Aggarwal M, McKenna R. Update on carbonic anhydrase inhibitors: a patent review (2008 - 2011). Expert Opin Ther Pat 2012;22:903–915.
- Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov 2008;7:168–181.
- Thiry A, Dogné JM, Masereel B, Supuran CT. Targeting tumor-associated carbonic anhydrase IX in cancer therapy. Trends Pharmacol Sci 2006;27:566–573.
- Brahimi-Horn MC, Pouysségur J. Oxygen, a source of life and stress. FEBS Lett 2007;581:3582–3591.
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 1999;399:271–275.
- Semenza GL. Hypoxia and cancer. Cancer Metastasis Rev 2007;26:223–224.
- Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW et al. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature 2002;417:975–978.
- Neri D, Supuran CT. Interfering with pH regulation in tumours as a therapeutic strategy. Nat Rev Drug Discov 2011;10:767–777.
- Aggarwal M, Kondeti B, McKenna R. Insights towards sulfonamide drug specificity in a-carbonic anhydrases. Bioorg Med Chem 2012 (In Press).
- Vullo D, De Luca V, Scozzafava A, Carginale V, Rossi M, Supuran CT et al. The first activation study of a bacterial carbonic anhydrase (CA). The thermostable a-CA from Sulfurihydrogenibium yellowstonense YO3AOP1 is highly activated by amino acids and amines. Bioorg Med Chem Lett 2012;22:6324–6327.
- De Luca V, Vullo D, Scozzafava A, Carginale V, Rossi M, Supuran CT et al. Anion inhibition studies of an a-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Bioorg Med Chem Lett 2012;22:5630–5634.
- Fulke AB, Mudliar SN, Yadav R, Shekh A, Srinivasan N, Ramanan R et al. Bio-mitigation of CO(2), calcite formation and simultaneous biodiesel precursors production using Chlorella sp. Bioresour Technol 2010;101:8473–8476.
- Ramanan R, Kannan K, Deshkar A, Yadav R, Chakrabarti T. Enhanced algal CO(2) sequestration through calcite deposition by Chlorella sp. and Spirulina platensis in a mini-raceway pond. Bioresour Technol 2010;101:2616–2622.
- Hunt JA, Lesburg CA, Christianson DW, Thompson RB, Fierke CA. Active-site engineering of carbonic anhydrase and its application to biosensors. EXS 2000:221–240.
- Kanbar B, Ozdemir E. Thermal stability of carbonic anhydrase immobilized within polyurethane foam. Biotechnol Prog 2010;26:1474–1480.
- Vinoba M, Lim KS, Lee SH, Jeong SK, Alagar M. Immobilization of human carbonic anhydrase on gold nanoparticles assembled onto amine/thiol-functionalized mesoporous SBA-15 for biomimetic sequestration of CO2. Langmuir 2011;27:6227–6234.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 2004;25:1605–1612.