Abstract
A series of phenols incorporating tertiary amine and trans-pyridylethenyl-carbonyl moieties were assayed as inhibitors of the β-carbonic anhydrase (CA, EC 4.2.1.1) from Saccharomyces cerevisiae, ScCA. One of these compounds was a low nanomolar ScCA inhibitor, whereas the remaining ones inhibited the enzyme with KIs in the range of 23.5–95.4 nM. The off-target human (h) isoforms hCA I and hCA II were much less inhibited by these phenols, with KIs in the range of 0.78–23.5 µM (hCA I) and 10.8–52.4 µM (hCA II). The model organism S. cerevisiae and this particular enzyme may be useful for detecting antifungals with a novel mechanism of action compared to the classical azole drugs to which significant drug resistance emerged.
Introduction
Fungi encode carbonic anhydrases (CAs, EC 4.2.1.1) belonging to at least two families, the α- and β-class such enzymesCitation1–5. Five distinct genetic families (α-, β-, γ-, δ- and ζ-CAs) encode such enzymes in organisms all over the phylogenetic treeCitation6–15. CAs efficiently catalyze the simple but essential reaction between carbon dioxide and water, with generation of bicarbonate and protons, which is too slow without a catalyst at neutral pH values, by using a metal hydroxide as nucleophile, at nearly neutral pH valuesCitation10–20. All chemical species involved in the CA-catalyzed reaction, carbon dioxide, bicarbonate and protons, are involved in many crucial physiologic processes and their perturbation may lead to dysfunctionsCitation10–20.
CAs also play important roles in fungi/yeasts, being involved in the CO2-sensing mechanism of these organisms and in their regulation of sexual developmentCitation1–5. In fact physiological concentrations of induce prominent virulence attributes in Candida albicans (e.g. filamentation) and capsule biosynthesis in Candida glabrata or Cryptococcus neoformans, through the direct activation of the fungal adenylyl cyclaseCitation2–6,Citation8.
equilibration by fungal CAs, both related to CO2 sensing and pathogenesisCitation2–6, were thoroughly investigated in several species. For example, the C. albicans enzyme CaNce103 or the orthologous C. glabrata one (CgNce103) – both belonging to the β-CA family – were shown to be essential for pathogenesis of these fungi in niches where CO2 availability is limited (e.g. the skin), or essential for the growth of C. neoformans in its natural environment (the enzyme from this pathogen was denominated Can2)Citation6. The newly discovered bZIP transcription factor CgRca1p is also required in such processes in various species such as C. albicans, C. glabrata and S. cerevisiaeCitation2–6,Citation8–13. Thus, the link between cAMP signaling and
sensing is conserved in fungi/yeasts and revealed CO2 sensing to be an important mediator of fungal metabolism and pathogenesisCitation2–6,Citation8–13.
The model organism Saccharomyces cerevisiae also encodes for a β-CA (the gene encoding this enzyme is denominated as Nce103), which is required for providing sufficient bicarbonate to essential metabolic carboxylation reactions for the yeast metabolism, such as those catalyzed by pyruvate carboxylase (PC), acetyl-CoA carboxylase (ACC), carbamoyl phosphate synthase (CPSase) and phosphoribosylaminoimidazole (AIR) carboxylaseCitation11–13. This enzyme (ScCA) has been investigated for its inhibition and activation with a range of modulators of activity such as sulfonamides and anions as inhibitors, and amines/amino acids as activatorsCitation11–13. Another yeast which has been investigated recently for the presence of CAs was Malassezia globosa, the organism causing dandruffCitation14. As other fungi/yeasts, M. globosa contains only one β-CA, which has been denominated as MgCACitation14. For this enzyme, a proof-of-concept study showed that inhibition of MgCA with sulfonamides has antifungal effects in vivo, in an animal model of dandruffCitation14, leading to the proposal of the fungal CAs from such pathogens as a possible antifungal target.
Apart from sulfonamides and their isosteres (sulfamates, sulfamides, etc) which represent the main class of CA inhibitors (CAIs)Citation16–26, CAs are also inhibited by phenols, through a different mechanism of actionCitation7. Indeed, whereas sulfonamides coordinate (in deprotonated state, as anions) to the metal ion from the enzyme active site, phenols (and several other classes of less investigated CAIs) anchor to the water molecule/hydroxide ion coordinated to the metal ionCitation7,Citation15,Citation21. Phenols were investigated in some detail for the inhibition of the many mammalian CA isoforms (CA I–XV)Citation27–29, but not such studies for the inhibition of fungal β-CAs are available so far. Here we report the first phenol inhibition study of a fungal/yeast β-CA, more precisely, we investigated the inhibition of ScCA with a series of phenols incorporating tertiary amine and pyridylethenyl-carbonyl moieties, recently reported by one of our groupsCitation30.
Materials and methods
Chemistry
Phenols 1–13 used in this study were reported earlier by one of our groupsCitation30. Acetazolamide used as standard was from Sigma-Aldrich (Milan, Italy). Buffers and inorganic reagents were of highest grade purity available from Sigma-Aldrich (Milan, Italy).
CA inhibition
An Applied Photophysics stopped-flow instrument has been used for assaying the CA catalyzed CO2 hydration activityCitation31. Phenol red (at a concentration of 0.2 mM) has been used as an indicator, working at the absorbance maximum of 557 nm, with 20 mM Hepes (pH 7.4 for hCA I and II) or with 20 mM Tris (pH 8.4, for ScCA) as buffers, and 20 mM NaClO4 (for maintaining constant the ionic strength), following the initial rates of the CA-catalyzed CO2 hydration reaction for a period of 10–100 s. The CO2 concentrations ranged from 1.7 to 17 mM for the determination of the kinetic parameters and inhibition constants. For each inhibitor at least six traces of the initial 5%–10% of the reaction have been used for determining the initial velocity. The uncatalyzed rates were determined in the same manner and subtracted from the total observed rates. Stock solutions of inhibitor (10 mM) were prepared in distilled-deionized water and dilutions up to 0.001 nM were done thereafter with the assay buffers. Inhibitor and enzyme solutions were preincubated together for 15 min at room temperature prior to assay, in order to allow for the formation of the E–I complex. The inhibition constants were obtained by non-linear least-squares methods using PRISM 3 (GraphPad Software, La Jolla, CA), as reported earlierCitation11–13, and represent the mean from at least three different determinations. All CA isoforms were recombinant ones obtained as reported earlier in our laboratoriesCitation11–13,Citation32,Citation33. The concentration of the enzymes in the assays was of 8.0 nM for hCA I, of 5.9 nM for hCA II and of 12.5 nM for ScCA, respectively.
Results and discussion
Inhibition data for two human CA isoforms (hCA I and II) and ScCA with compounds 1–13 and acetazolamide (AAZ, 5-acetamido-1,3,4-thiadiazole-2-sulfonamide, as standard inhibitor) are shown in .
Table 1. Human (h) hCA I, II and yeast (S. cerevisiae) ScCA inhibition data with phenols 1–13 by a stopped flow CO2 hydrase assayCitation31.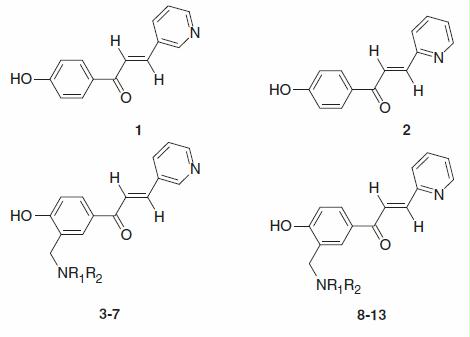
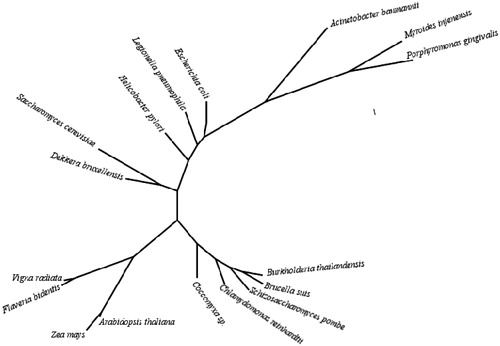
shows an alignment of the amino acid sequences of selected β-CAs from fungal, bacterial, plant and algal species, whereas present a phylogenetic analysis of many such enzymes from a large variety of bacteria, fungi/yeasts, algae and plants.
Figure 1. Alignment of the amino acid sequences of selected β-CAs from four species. The S. cerevisiae numbering system was used. Zinc ligands (amino acids: 57, 112 and 115) are indicated in red whereas the catalytic dyad (amino acids 59 and 61) typical of β-CAs in blue. The asterisk (*) indicates identity at a position; the symbol (:) designates conserved substitutions, while (·) indicates semi-conserved substitutions. Multiple alignment was performed with the program Clustal W, version 2.1 (http://www.ebi.ac.uk/services/proteins). Sequence accession numbers are indicated in .

Figure 2. Phylogenetic tree of the beta CAs from selected eukaryotic and prokaryotic species. The tree was constructed using the program PhyML 3.0 (http://www.atgc-montpellier.fr/phyml/). Organisms and sequence accession numbers are as follows: Schizosaccharomyces pombe (CAA21790); Brucella suis 1330 (NP_699962.19); Burkholderia thailandensis Bt4 (ZP_02386321); Coccomyxa sp. (AAC33484.1); Chlamydomonas reinhardtii (XP_001699151.1); Acinetobacter baumannii (YP_002326524); Porphyromonas gingivalis ATCC 33277 (YP_001929649.1); Myroides injenensis (ZP_10784819); Zea mays (NP_001147846.1); Vigna radiata (AAD27876); Flaveria bidentis, isoform I (AAA86939.2); Arabidopsis thaliana (AAA50156); Helicobacter pylori (BAF34127.1); Legionella pneumophila 2300/99 (YP_003619232); Escherichia coli (ACI70660); Saccharomyces cerevisiae (GAA26059); Dekkera bruxellensis AWRI1499 (EIF49256).
Data from show that the human (off-target) isoforms are poorly inhibited by phenols 1–13, investigated here, which typically had inhibition constants in the micromolar range. For example, against hCA I the KIs were in the range of 0.78–23.5 µM, whereas against hCA II in the range of 10.8–2.4 µM. The unsubstituted phenols 1 and 2 were among the most effective hCA I inhibitors (KIs were in the range of 0.78–1.77 µM), and the introduction of tertiary amine moieties in ortho generally leads them to a strong diminution of the inhibitory activity. This is even more evident in the case of hCA II, for which the simple phenols 1 and 2 were again the best inhibitors (KIs were in the range of 10.8–12.5 µM), whereas the tertiary amines 3–13 were generally much weaker inhibitors (). It should be observed that the sulfonamide AAZ is a much more potent inhibitor of both these α-CAs, with KIs in the range of 12–250 nM.
Interestingly, the phenols 1–13 investigated here showed very good ScCA inhibitory activity, with KIs in the range of 4.9–95.4 nM (). This is an unexpected result due to the fact that phenols were generally weaker inhibitors against most investigated CAsCitation3,Citation4, but it should be mentioned, as noted in the “Introduction” section, that phenols were not yet investigated as ScCA inhibitors till the present work. The two parent compounds 1 and 2 were medium potency ScCA inhibitors, with inhibition constants in the range of 65.8–76.5 nM, similar to the sulfonamide AAZ (KI of 82.6 nM, investigated earlier by these groups)Citation11. However, many of the derivatized phenols 3–13 showed enhanced ScCA inhibitory action compared to the parent phenols 1 and 2. For example, compound 5, incorporating the 3-pyridiyl moiety and the pyrrolidine ring in ortho to the phenol moiety, was a low nanomolar (KI of 4.9 nM) inhibitor of the yeast enzyme. Interestingly, its homolog 6, with an extra CH2 moiety (piperidine instead of pyrrolidine ring) was 13.2 times a weaker ScCA inhibitor compared to 5. Thus, small structural changes of the scaffold lead to dramatic changes of the ScCA inhibitory action in this class of compounds. Structure–activity relationship (SAR) is in fact rather complicated. The nature of the R1 and R2 groups substituting the amine functionality seem to be the major factor influencing enzyme inhibitory activity. However, this influence is rather complicated. For example, for the 3-pyridyl-substituted compounds, activity is increasing with the introduction of the Me2N moiety in the lead 1 (compound 3, KI of 23.5 nM) but then decrease again for the diethylamino derivative 4 (KI of 77.1 nM). As mentioned above, the cyclic derivative 5 is the most potent ScCA inhibitor, but then its homolog 6 and the similar compound 7 showed activities comparable to that of the diethylamino derivative 4. For the 2-pyridyl-substituted series activity of the tertiary amines 8–13 was lower compared to that of the lead 2, making this subseries less interesting.
But how can these results be rationalized, considering than no X-ray crystal structure of ScCA in adducts with inhibitors is available? In order to explain these data, we aligned the amino acid sequences of the ScCA enzyme with those of other β-CAs investigated earlier, from bacterial plant or algae ().
As seen from and , ScCA shares many common feature with the corresponding β-CAs from other fungi/yeasts, bacteria or plants/algae. As seen from the alignment of , all these enzymes possess the typical β-CA zinc-coordinating ligands, i.e. two cysteine and one histidine residue, in the specific case Cys57, His112 and Cys115 (ScCA numbering system is used throughout this paperCitation11–13). The fourth zinc ligand is a water molecule/hydroxide ion which acts as nucleophile in the catalytic cycle, attacking the CO2 molecule bound in a hydrophobic pocket nearby. However, unlike the highly investigated α-class CAs, in which the zinc-bound hydroxide is easily generated (and it participates in a hydrogen bond network with Thr199 and Asp106), for the β-CAs a different catalytic dyad participates in the activation of the zinc-coordinated water molecule, constituted by Asp59 and Arg61 (), which is also present in ScCA and conserved in all β-CAs investigated so farCitation11–13.
Furthermore, the phylogenetic tree from , in which all fungal/yeast β-CAs for which the sequence is available in data bases were included, shows these enzymes to be related with each other, all of them clustering on very nearby branches of the tree. It is interesting to note that the enzyme more phylogenetically similar to ScCA is the one from Dekkera bruxellensis (DbrCA). These enzymes cluster on a branch, which is intermediate bewteen the bacterial β-CAs and such plant/algal enzymes. However, the yeast S. pombe did not cluster with the other yeast but on a separate branch together with some bacterial enzymes (from B. thailandensis and B. suis) ().
Conclusions
In conclusion, we report here a series of phenols incorporating tertiary amine and trans-pyridylethenyl-carbonyl moieties, which were assayed as inhibitors of the β-carbonic anhydrase (CA, EC 4.2.1.1) from S. cerevisiae, ScCA. One of these compounds was a low nanomolar ScCA inhibitor (KI of 4.9 nM), whereas the remaining ones inhibited the enzyme with KIs in the range of 23.5–95.4 nM. The off-target human (h) isoforms hCA I and hCA II were much less inhibited by these phenols, with KIs in the range of 0.78–23.5 µM (hCA I) and 10.8–52.4 µM (hCA II). The model organism S. cerevisiae and this particular enzyme may be useful for detecting antifungals with a novel mechanism of action compared to the classical azole drugs to which significant drug resistance emerged, also considering the fact that we have recently shown that some potent sulfonamide ScCA inhibitors also inhibit the growth of the yeast in vivoCitation34.
Declaration of interest
The authors declare no conflict of interest.
This research was financed by two 7th FP EU projects (Metoxia and Dynano). Semra Isik was supported by a grant from the Council of Higher Education of Turkey, being on leave from Balikesir University, Turkey.
References
- Elleuche S, Pöggeler S. Carbonic anhydrases in fungi. Microbiology 2010;156:23–9
- Klengel T, Liang WJ, Chaloupka J, et al. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol 2005;15:2021–6
- Hall RA, Mühlschlegel FA. Fungal and nematode carbonic anhydrases: their inhibition in drug design. In: Supuran CT, Winum JY, eds. Drug design of zinc-enzyme inhibitors: functional, structural, and disease applications. Hoboken: John Wiley & Sons; 2009:301–22
- Ohndorf UM, Schlicker C, Steegborn C. Crystallographic studies on carbonic anhydrases from fungal pathogens for structure-assisted drug development. In: Supuran CT, Winum JY, eds. Drug design of zinc-enzyme inhibitors: functional, structural, and disease applications. Hoboken: John Wiley & Sons; 2009:323–34
- Innocenti A, Leewattanapasuk W, Mühlschlegel FA, et al. Carbonic anhydrase inhibitors. Inhibition of the β-class enzyme from the pathogenic yeast Candida glabrata with anions. Bioorg Med Chem Lett 2009;19:4802–5
- Schlicker C, Hall RA, Vullo D, et al. Structure and inhibition of the CO2-sensing carbonic anhydrase Can2 from the pathogenic fungus Cryptococcus neoformans. J Mol Biol 2009;385:1207–20
- De Simone G, Supuran CT. (In)organic anions as carbonic anhydrase inhibitors. J Inorg Biochem 2012;111:117–29
- Cottier F, Leewattanapasuk W, Kemp LR, et al. Carbonic anhydrase regulation and CO2 sensing in the fungal pathogen Candida glabrata involves a novel Rca1p ortholog. Bioorg Med Chem 2013;21:1549–54
- Carta F, Innocenti A, Hall RA, et al. Carbonic anhydrase inhibitors. Inhibition of the β-class enzymes from the fungal pathogens Candida albicans and Cryptococcus neoformans with branched aliphatic-/aromatic carboxylates and their derivatives. Bioorg Med Chem Lett 2011;21:2521–6
- Innocenti A, Mühlschlegel FA, Hall RA, et al. Carbonic anhydrase inhibitors. Inhibition of the beta-class enzymes from the fungal pathogens Candida albicans and Cryptococcus neoformans with simple anions. Bioorg Med Chem Lett 2008;18:5066–70
- Isik S, Kockar F, Aydin M, et al. Carbonic anhydrase inhibitors. Inhibition of the β-class enzyme from the yeast Saccharomyces cerevisiae with sulfonamides and sulfamates. Bioorg Med Chem 2009;17:1158–63
- Isik S, Kockar F, Arslan O, et al. Carbonic anhydrase inhibitors. Inhibition of the β-class enzyme from the yeast Saccharomyces cerevisiae with anions. Bioorg Med Chem Lett 2008;18:6327–31
- Isik S, Kockar F, Aydin M, et al. Carbonic anhydrase activators: activation of the β-carbonic anhydrase Nce103 from the yeast Saccharomyces cerevisiae with amines and amino acids. Bioorg Med Chem Lett 2009;19:1662–5
- Hewitson KS, Vullo D, Scozzafava A, et al. Molecular cloning, characterization and inhibition studies of a β-carbonic anhydrase from Malassezia globosa, a potential antidandruff target. J Med Chem 2012;55:3513–20
- Alterio V, Di Fiore A, D’Ambrosio K, et al. Multiple binding modes of inhibitors to carbonic anhydrases: how to design specific drugs targeting 15 different isoforms? Chem Rev 2012;112:4421–68
- Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov 2008;7:168–81
- Supuran CT. Structure-based drug discovery of carbonic anhydrase inhibitors. J Enzyme Inhib Med Chem 2012;27:759–72
- Supuran CT. Carbonic anhydrases: from biomedical applications of the inhibitors and activators to biotechnologic use for CO2 capture. J. Enzyme Inhib Med Chem 2013;28:229–30
- Supuran CT. Carbonic anhydrase inhibitors. Bioorg Med Chem Lett 2010;20:3467–74
- Singh S, Supuran CT. QSARs on human carbonic anhydrase VA and VB inhibitors of some new not yet synthesized, substituted aromatic/heterocyclic sulphonamides as anti-obesity agent. J Enzyme Inhib Med Chem 2012;27:666–72
- Ekinci D, Kurbanoglu NI, Salamcı E, et al. Carbonic anhydrase inhibitors: inhibition of human and bovine isoenzymes by benzenesulphonamides, cyclitols and phenolic compounds. J Enzyme Inhib Med Chem 2012;27:845–8
- Bootorabi F, Jänis J, Hytönen VP, et al. Acetaldehyde-derived modifications on cytosolic human carbonic anhydrases. J Enzyme Inhib Med Chem 2011;26:862–70
- Supuran CT. Bacterial carbonic anhydrases as drug targets: toward novel antibiotics? Front Pharmacol 2011;2:1--6
- Supuran CT, Scozzafava A, Casini A. Carbonic anhydrase inhibitors. Med Res Rev 2003;23:146–89
- Neri D, Supuran CT. Interfering with pH regulation in tumours as a therapeutic strategy. Nature Rev Drug Discov 2011;10:767–77
- Liu F, Martin-Mingot A, Lecornué F, et al. Carbonic anhydrases inhibitory effects of new benzenesulfonamides synthesized by using superacid chemistry. J Enzyme Inhib Med Chem 2012;27:886–91
- Innocenti A, Vullo D, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors. Interactions of phenols with the 12 catalytically active mammalian isoforms (CA I – XIV). Bioorg Med Chem Lett 2008;18:1583–7
- Innocenti A, Vullo D, Scozzafava A, Supuran CT. Carbonic anhydrase inhibitors. Inhibition of mammalian isoforms I–XIV with a series of substituted phenols including paracetamol and salicylic acid. Bioorg Med Chem 2008;16:7424–8
- Sahin H, Aliyazicioglu R, Yildiz O, et al. Honey, pollen, and propolis extracts show potent inhibitory activity against the zinc metalloenzyme carbonic anhydrase. J Enzyme Inhib Med Chem 2011;26:440–4
- Bilginer S, Gul HI, Mete E, et al. 1-(3-Aminomethyl-4-hydroxyphenyl)-3-pyridinyl-2-propen-1-ones: a novel group of tumour-elective cytotoxins. J Enzyme Inhib Med Chem 2013;28:1--7
- Khalifah RG. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J Biol Chem 1971;246:2561–73
- Kolayli S, Karahalil F, Sahin H, et al. Characterization and inhibition studies of an α-carbonic anhydrase from the endangered sturgeon species Acipenser gueldenstaedti. J Enzyme Inhib Med Chem 2011;26:895–900
- Ozensoy O, Arslan M, Supuran CT. Carbonic anhydrase inhibitors: purification and inhibition studies of pigeon (Columba livia var. domestica) red blood cell carbonic anhydrase with sulfonamides. J Enzyme Inhib Med Chem 2011;26:749–53
- Alafeefy AM, Isik S, Al-Jaber NA, et al. Carbonic anhydrase inhibitors. Benzenesulfonamides incorporating cyanoacrylamide moieties strongly inhibit Saccharomyces cerevisiae β-carbonic anhydrase. Bioorg Med Chem Lett 2013;23:3570--5

