Abstract
All proteolytic enzymes, which are able to renature and reacquire the proteolytic activity on a copolymerized substrate, can be analyzed by zymography upon removal of sodium dodecyl sulfate (SDS). Protonography, the new technique described in this study, unlike zymography, allows the detection of a different protein, not a protease, i.e. of the carbonic anhydrase (CA, EC 4.2.1.1) activity on a SDS polyacrylamide gel electrophoresis gel. CAs are zinc-containing enzymes that catalyze the reversible conversion of carbon dioxide to bicarbonate and protons. Hydrogen ions produced during the catalyzed reaction are responsible for the change of color that appears on the gel around the CA band. For this reason, we named the new technique “protonography”. The following four salient features characterize this new technique: (a) on the basis of molecular weight markers, recombinant or native CAs with different molecular weights can be detected and quantified rapidly on a single gel; (b) the hydratase activity can be reversibly inhibited by SDS during electrophoresis and recovered by incubating the gel in aqueous Triton X-100; (c) it is possible to separate active oligomeric forms of CAs on the gel enabling their activities to be determined independently of one another. This feature is not possible when using solution assays; and (d) it can be a useful tool to establish if a putative or a newly identified CA in a genome is expressed and enzymatically active. This article outlines the general principles employed in protonography, providing an easy procedure to implement it in laboratories working with CAs. It also presents an overview of its development and current research applications through specific examples.
Introduction
In 1978, a new technique was developed to study plasminogen activators, proteins present in the blood and involved in the coagulation cascade; this technique was named “in gel zymography”Citation1. It used a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel to separate plasmin (produced by the proteolysis of plasminogen) from other molecules. Plasmin detection was based on the degradation of fibrin (which is one of the substrates of this protease) contained in an agarose gel, named indicator gelCitation1,Citation2. Enzyme diffusion into the indicator gel occurred during incubation of the gel sandwich and resulted in degradation of the protein substrate. The indicator gel was then stained to reveal zones of proteolysis, which appeared as clear bands on a dark background. Subsequently, the method was optimized by incorporating the protein substrate directly into the SDS-PAGE gelCitation2,Citation3. Thus, as aforementioned, zymography is defined as a simple electrophoretic method and functional assay for measuring proteolytic activity and is widely applied for investigating proteases belonging to all classes (serine-/threonine-, cysteine, aspartic and metalloproteases)Citation2,Citation3. The standard method is based on SDS gels impregnated with a protein substrate, in particular gelatin or casein. All the proteolytic enzymes, which are able to renature and reacquire the proteolytic activity on a copolymerized substrate, upon removal of SDS (acting as a denaturant), can be analyzed by this technique. As described in many literature reports, zymography offers several advantages with respect to other methods because it is feasible with inexpensive materials, and it is also a very sensitive technique, with detection limits of about 10 pg of protease on gelatin zymogram, when compared with other methods, such as ELISACitation2–4.
In this article, we introduce a new technique, similar to zymography and derived from it, and named in this study by our group, “protonography”. Unlike zymography, this new technique allows the detection of the activity of a different class of proteins, the carbonic anhydrase (CA, EC 4.2.1.1) on a SDS-PAGE gel. In the past, Kotwica et al. used a PAGE zymography without SDS to detect the presence of CA activity in the upper vas deferens of the cotton leaf worm, Spodoptera littoralisCitation5. However, this zymography-like technique applied to CAs did not allow determining of the molecular weight of the enzymes. Moreover, Drescher reported the use of a fluorescent CA inhibitor, 5-dimethylaminonaphthalene-1-sulfonamide, to identify the CA on polyacrylamide gel by an imaging system apparatusCitation6,Citation7.
CAs catalyze a simple reaction important in all life kingdoms: the carbon dioxide hydration to bicarbonate and protons: CO2 + H2O ⇔ + H+Citation8–18. The name “protonography” was chosen because CA activity produces hydrogen ions (protons) during the catalyzed reaction, which are then responsible of the change of color that appears on the gel, in correspondence with a CA, CO2 hydrase activity band. The band of the protein with CA activity becomes yellow (see later in the text). The advantage of this technique is that on the basis of molecular weight markers, recombinant or native CAs with different molecular weights can be detected and quantified rapidly on a single gel.
Five different, genetically distinct CA families are known to date, the α-, β-, γ-, δ- and ζ-CAsCitation10,Citation11,Citation13,Citation19–26. Whereas α-, β- and δ-CAs use Zn(II) ions at the active site; the γ-CAs are probably Fe(II) enzymesCitation11,Citation19,Citation27–30 (but they are active also with bound Zn(II) or Co(II) ions)Citation30, whereas the ζ-class uses Cd(II) or Zn(II) to perform the physiologic reactionCitation10–14,Citation19–26,Citation31–34. The molecular weight of these CA classes is in the range of 20–30 kDa on an SDS-PAGE gelCitation8–11,Citation13,Citation14,Citation22–27,Citation35–49.
In this study, we present the protonography as a new technique for the rapid detection of CA activity, exemplifying it on one α-CA belonging to the mammalian organism Bos taurus (bovine α-CA; bCA), and one α-CA from the pathogenic bacterium, Vibrio cholerae (VchCA).
Materials and methods
Mammalian bCA
bCA was supplied by Sigma-Aldrich (St. Louis, MO) as crude extract from bovine erythrocytes (lyophilized powder).
Construct preparation, protein expression and purification of VchCA
The GeneArt Company, specialized in gene synthesis, designed the synthetic V. cholerae gene encoding for the α-CA, lacking the signal peptide (i.e. the first 20 amino acid residues of the peptide sequence) and containing a NdeI and XhoI site at the 5′ and 3′ end of the VchCA gene, respectively. The resulting plasmid was amplified into Escherichia coli DH5 α cells. The V. cholerae DNA fragments were separated on 1% agarose gel. The recovered V. cholerae gene and the linearized expression vector (pET15-b) were ligated by T4 DNA ligase to form the expression vector pET15-b/Vch. In order to confirm the integrity of the V. cholerae gene and that no errors occurred at the ligation sites, the pET15-b/Vch vector was sequenced. Competent E. coli BL21 (DE3) cells were transformed with pET15-b/Vch vector, induced with 1 mM IPTG and grown at 37 °C for 5 h. After additional growth for 5 h, cells were harvested and disrupted by sonication at 4 °C. Following centrifugation, the cell extract was placed in conical flasks, and an amount of ammonium sulfate gradually added to obtain saturations of 45%. After resting for 14 h at 4 °C, the sample was centrifuged at 12 000 × g at 4 °C for 30 min. The precipitate was resuspended in 20 mM buffer phosphate, pH 8.0, and loaded onto a His-select HF Nickel affinity gel. The protein was eluted with 250 mM imidazole. At this stage of purification, the enzyme was at least 95% pure, and the obtained recovery was of 30 mg of the recombinant bacterial CA.
CA assay
An Applied Photophysics stopped-flow instrument was used for assaying the CA catalyzed CO2 hydration activityCitation50. Phenol red (at 0.2 mM) was used as indicator, working at the absorbance maximum of 557 nm with 10 mM HEPES (pH 7.5) as buffer and 0.1 M NaClO4 (for maintaining constant ionic strength), at 20 °C, following the CA-catalyzed CO2 hydration reaction for a period of 10–100 s (the uncatalyzed reaction needs around 60–100 s in the assay conditions, whereas the catalyzed ones are of around 6–10 s). The CO2 concentrations ranged from 1.7 to 17 mM for the determination of the kinetic parameters. The uncatalyzed rates were determined in the same manner and subtracted from the total observed rates. Enzyme concentrations in the assay system were 9.1 nM for bCA and 8.6 nM for VchCA.
Protonography
SDS-PAGE was performed as described by LaemmliCitation51. Wells of 12% SDS gel were loaded with bCA or VchCA mixed with Laemmli loading buffer without 2-mercaptoethanol and without boiling the samples, in order to avoid protein denaturation. The gel was run at 180 V until the dye front ran off the gel. Following the electrophoresis, the gel was removed from glass plates and soaked in 2.5% Triton X-100 for 1 h on a shaker and then twice in 100 mM Tris, pH 8.2 containing 10% isopropanol for 10 min. Subsequently, the gel was incubated in 0.1% bromothymol blue in 100 mM Tris, pH 8.2 for 30 min and then immersed in CO2-saturated ddH2O to visualize the hydratase activity of the enzyme. The assay was performed at room temperature, and the CO2-satured solution was prepared by bubbling CO2 into 200 mL distilled water for approximately 3 h. The local decrease in pH due to the presence of CA activity was evidenced by the formation of yellow bands due to the change of the indicator color from blue (alkaline pH) to yellow (acidic pH).
Preparation of cell lysates from E. coli
We used E. coli BL21 (DE3)pLysS cells from Agilent Technologies. Cells were grown for 16 h at 37 °C, then harvested and disrupted by sonication at 4 °C. Following centrifugation, the extract was assayed using the Bradford assay. A total of 30 µg of protein per well was loaded on a 12% SDS-PAGE without 2-mercaptoethanol and without boiling the sample. Gels were made in duplicate. One was stained with Coomassie, and the other was subject to the protonography technique as described in this section.
Results and discussion
The recombinant VchCA prepared as described in “Materials and methods” was isolated and purified to homogeneity at room temperature from E. coli (DE3) cell extracts. CA activity was recovered in the soluble fraction of the cell extract obtained after sonication and centrifugation. By use of the affinity column (His-select HF nickel affinity gel), VchCA was purified to about 95% of purity.
A stopped-flow CO2 hydrase assay has been used to measure the catalytic activity of VchCA and bCA in identical conditions. shows the catalytic activity of these enzymes for the CO2 hydration reaction. It may be observed that VchCA has kinetic parameters quite similar to those of the bCA.
Table 1. Kinetic parameters for CO2 hydration reaction catalyzed by bCA and VchCA at 20 °C and pH 7.5. Ki for acetazolamide was 10 nM for bCA and 6.8 nM for VchCA.
To investigate the hydratase activity of VchCA and bCA on the SDS-PAGE gel, samples of VchCA or bCA were prepared diluting a stock solution and loaded on the gel in the range of 100–1000 ng/well. As described in the experimental section, the protonography technique is based on the monitoring of pH variation in the gel (protonogram) due to the catalyzed conversion of CO2 to bicarbonate and protons. The protonogram was stained with bromothymol blue, which is an indicator of pH variation. This dye appears blue in its deprotonated form, while its color changes to yellow in the protonated form. Thus, the production of hydrogen ions during the CO2 hydration reaction, due to the bCA or VchCA hydratase activity lowers the pH of the solution until the color transition point of the dye is reached (pH 6.8). and show a protonogram obtained with different concentration of VchCA and bCA ranging from 10 to 1400 ng. The yellow band corresponded to the CA position on the gel. The time required for the color change is inversely related to the quantity of CA, which is responsible for dropping the pH from 8.2 to the transition point of the dye in a control buffer. The success of this technique is that the hydratase activity can be reversibly inhibited by SDS during electrophoresis and recovered by incubating the gel in aqueous Triton X-100.
Figure 1. Protonogram obtained at different concentrations of bCA. The yellow band corresponds to the bCA position on the gel responsible for the drop of pH from 8.2 to the transition point of the dye in the control buffer. Three different incubation times are used (5, 10 and 15 s).
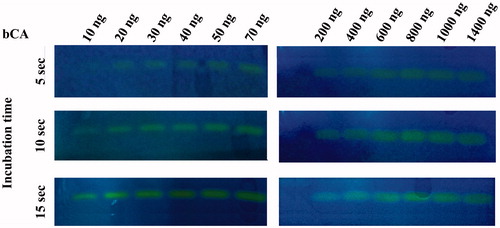
Figure 2. Protonogram obtained at different concentrations of VchCA. The yellow band corresponds to the VchCA position on the gel. Incubation times were 10, 20 and 30 s, higher than those indicated for bCA and shown in .
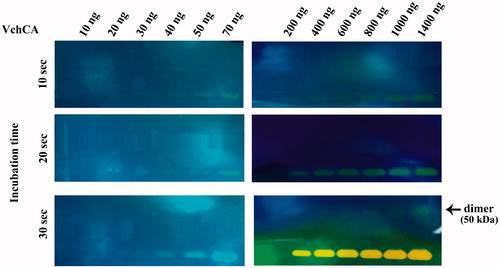
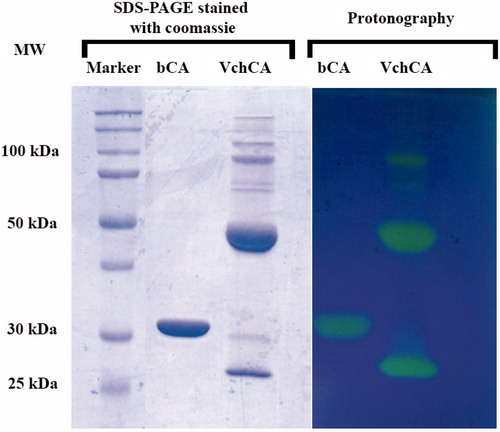
shows a comparison between the protonography and SDS-PAGE stained with Coomassie blue. Both gels were run under denaturing but non-reducing conditions. After the electrophoretic separation of proteins, the gel on the left-hand of was stained with Coomassie blue, whereas the gel on the right-hand of the same figure was resolved by exchange of SDS with a nonionic detergent such as Triton X-100 and then stained with bromothymol blue. The CA activity was detected by immersing the gel in CO2-saturated ddH2O. The enzyme activity was detected as yellow bands against the blue background. Moreover, shows that protonography allowed us to identify different oligomeric states of the VchCA when the gel is overloaded (4000 ng of protein per well). This protein, in fact, is a monomer of 25 kDa but the protonogram showed three visible bands corresponding to the dimeric (50 kDa) and tetrameric (100 kDa) forms. At high concentration of protein loaded per well, in the case of bCA, it is possible to see that the protein is always monomeric, with a molecular weight of 30 kDa, whereas for VchCA, it was possible to separate the different oligomeric forms of the enzymes, enabling their activities to be determined independently from one another, which is not possible in the solution assay. Thus, a specific advantage of this system is that the active, oligomeric forms of the enzyme, which can be distinguished on the basis of molecular weight, can be detected by this new technique. As shown in , VchCA already showed a band at the position of the dimeric state (50 kDa) in the well containing 1400 ng of enzyme.
Figure 3. Comparison between the protonography and SDS-PAGE stained with Coomassie blue. Both gels were run under denaturing but non-reducing conditions. Gels were loaded with 4000 ng/well of bCA and VchCA, respectively. The incubation time in this case was 5 s.
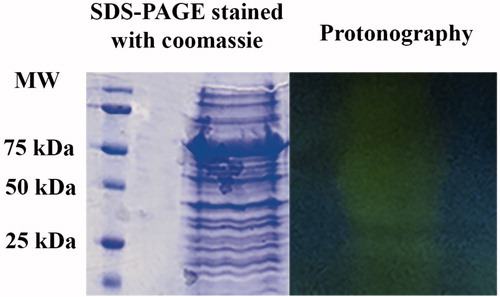
Another advantage of this technique is the possibility to reveal CAs in biological systems, such as tissues or bacterial extracts from which only the genome information is so far available. In this case, protonography might be the method of choice for CAs screening, identification and characterization. For example, we prepared a cellular extract of E. coli. The genome of E. coli encodes for two CA classes: β and γ-CAs, as described in the literatureCitation52,Citation53. In this study, we want to demonstrate that protonography allowed us to detect hydratase activity in the cellular extract of E. coli. shows a SDS-PAGE gel stained with Coomassie blue and a protonogram obtained after loading the bacterial extract on the gels. As expected, when analyzed by protonography, the whole cellular extract of E. coli revealed two main yellow bands at a molecular weight ranging from 25 to 50 kDa, corresponding to a monomeric (25 kDa) or dimeric (50 kDa) β-CA. It was difficult to evidence the band at 75 kDa corresponding to a trimeric γ-CA or, probably, it migrated under these experimental conditions as a monomer with a molecular weight of about 25 kDa. This is an interesting result in the study of organisms with known genome sequences for which we can identify putative CA by bioinformatics analysis, but we cannot ascertained if the ORFs are indeed expressed and active. Protonography can be a useful tool to help with this issue.
Figure 4. SDS-PAGE protonography using the whole cellular extract prepared from Escherichia coli cells. Both gels were run under denaturing but non-reducing conditions. Protonography shows two main yellow bands at a molecular weight ranging from 25 to 50 kDa (see text). Protonography thus evidenced an E. coli CA activity as described in the literature.
Furthermore, protonography coupled with mass spectrometry for protein identification will make possible the broader mapping of active CAs present in protein extract from various sources, allowing the detection of distinct CA isoforms.
Conclusions
Protonography is a new, cheap, reproducible and easy to perform technique for assaying CO2 hydratase activity on a SDS-PAGE gel. In this study, we presented the basic features of this new technique by using three examples of CAs belonging to a mammal (bCA), a bacterium (V. cholerae α-CA) and a bacterium for which the two CAs present in its genome (one β- and one γ-CA), were not investigated in great detail, i.e. E. coli. For all these three diverse examples, we were able to demonstrate the CO2 hydratase activity by using protonography, allowing us to speculate that this technique may be extended also to enzymes belonging to other CA classes than the α one. Indeed, combining protonography with mass spectrometric techniques could be particularly advantageous for the biochemical identification and analysis of putative, new CAs. Furthermore, this technique might reveal the expression of active CAs that has important roles in tumor progression and metastasis, such as CA II, CA IX and XII. In fact, there are many cases of tumors overexpressing such isoformsCitation54,Citation55. However, as outlined above, protonography may easily reveal whether CA-like proteins not yet characterized but present in the genome of many organisms, do possess enzymatic activity without the need to clone, overexpress and purify the protein, as exemplified by the E. coli experiments reported in this article.
Declaration of interest
The authors declare no conflict of interest.
References
- Granelli-Piperno A, Reich E. A study of proteases and protease-inhibitor complexes in biological fluids. J Exp Med 1978;148:223–34
- Vandooren J, Geurts N, Martens E, et al. Zymography methods for visualizing hydrolytic enzymes. Nat Methods 2013;10:211–20
- Leber TM, Balkwill FR. Zymography: a single-step staining method for quantitation of proteolytic activity on substrate gels. Anal Biochem 1997;249:24–8
- Kurschat P, Wickenhauser C, Groth W, et al. Identification of activated matrix metalloproteinase-2 (MMP-2) as the main gelatinolytic enzyme in malignant melanoma by in situ zymography. J Pathol 2002;197:179–87
- Kotwica J, Ciuk MA, Joachimiak E, et al. Carbonic anhydrase activity in the vas deferens of the cotton leafworm – Spodoptera littoralis (Lepidoptera: Noctuidae) controlled by circadian clock. J Physiol Pharmacol 2006;57:107–23
- Drescher DG. Purification of a carbonic anhydrase from the inner ear of the guinea pig. Proc Natl Acad Sci USA 1977;74:892–6
- Drescher DG. Purification of blood carbonic anhydrases and specific detection of carbonic anhydrase isoenzymes on polyacrylamide gels with 5-dimethylaminonaphthalene-1-sulfonamide (DNSA). Anal Biochem 1978;90:349–58
- Capasso C, Supuran CT. Anti-infective carbonic anhydrase inhibitors: a patent and literature review. Expert Opin Ther Pat 2013;23:693–704
- Capasso C, Supuran CT. Sulfa and trimethoprim-like drugs – antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J Enzyme Inhib Med Chem 2013. [Epub ahead of print]. doi: 10.3109/14756366.2013.787422
- De Luca V, Vullo D, Scozzafava A, et al. Anion inhibition studies of an alpha-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Bioorg Med Chem Lett 2012;22:5630–4
- Del Prete S, De Luca V, Vullo D, et al. Biochemical characterization of the gamma-carbonic anhydrase from the oral pathogen Porphyromonas gingivalis, PgiCA. J Enzyme Inhib Med Chem 2013. [Epub ahead of print] doi: 10.3109/14756366.2013.822371
- Del Prete S, Vullo D, De Luca V, et al. Biochemical characterization of the delta-carbonic anhydrase from the marine diatom Thalassiosira weissflogii, TweCA. J Enzyme Inhib Med Chem 2014. [Epub ahead of print]. doi: 10.3109/14756366.2013.868599
- Luca VD, Vullo D, Scozzafava A, et al. An alpha-carbonic anhydrase from the thermophilic bacterium Sulphurihydrogenibium azorense is the fastest enzyme known for the CO2 hydration reaction. Bioorg Med Chem 2013;21:1465–9
- Migliardini F, De Luca V, Carginale V, et al. Biomimetic CO2 capture using a highly thermostable bacterial alpha-carbonic anhydrase immobilized on a polyurethane foam. J Enzyme Inhib Med Chem 2014;29:146–50
- Supuran CT, Scozzafava A. Carbonic anhydrases as targets for medicinal chemistry. Bioorg Med Chem 2007;15:4336–50
- Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov 2008;7:168–81
- Supuran CT. Carbonic anhydrases – an overview. Curr Pharm Des 2008;14:603–14
- Supuran CT. Carbonic anhydrase inhibitors. Bioorg Med Chem Lett 2010;20:3467–74
- Vullo D, Del Prete S, Osman SM, et al. Sulfonamide inhibition studies of the gamma-carbonic anhydrase from the oral pathogen Porphyromonas gingivalis. Bioorg Med Chem Lett 2014;24:240–4
- Vullo D, Del Prete S, Osman SM, et al. Sulfonamide inhibition studies of the delta-carbonic anhydrase from the diatom Thalassiosira weissflogii. Bioorg Med Chem Lett 2014;24:275–9
- Nishimori I, Vullo D, Minakuchi T, et al. Anion inhibition studies of two new beta-carbonic anhydrases from the bacterial pathogen Legionella pneumophila. Bioorg Med Chem Lett 2014;24:1127–32
- Del Prete S, De Luca V, Scozzafava A, et al. Biochemical properties of a new alpha-carbonic anhydrase from the human pathogenic bacterium, Vibrio cholerae. J Enzyme Inhib Med Chem 2014;29:23–7
- Vullo D, Sai Kumar RS, Scozzafava A, et al. Anion inhibition studies of a beta-carbonic anhydrase from Clostridium perfringens. Bioorg Med Chem Lett 2013;23:6706–10
- Vullo D, De Luca V, Scozzafava A, et al. The extremo-alpha-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium azorense is highly inhibited by sulfonamides. Bioorg Med Chem 2013;21:4521–5
- Di Fiore A, Capasso C, De Luca V, et al. X-ray structure of the first ‘extremo-alpha-carbonic anhydrase', a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr D Biol Crystallogr 2013;69:1150–9
- Del Prete S, Isik S, Vullo D, et al. DNA cloning, characterization, and inhibition studies of an alpha-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. J Med Chem 2012;55:10742–8
- Del Prete S, Vullo D, De Luca V, et al. A highly catalytically active gamma-carbonic anhydrase from the pathogenic anaerobe Porphyromonas gingivalis and its inhibition profile with anions and small molecules. Bioorg Med Chem Lett 2013;23:4067–71
- Innocenti A, Zimmerman SA, Scozzafava A, et al. Carbonic anhydrase activators: activation of the archaeal beta-class (Cab) and gamma-class (Cam) carbonic anhydrases with amino acids and amines. Bioorg Med Chem Lett 2008;18:6194–8
- Zimmerman S, Innocenti A, Casini A, et al. Carbonic anhydrase inhibitors. Inhibition of the prokariotic beta and gamma-class enzymes from Archaea with sulfonamides. Bioorg Med Chem Lett 2004;14:6001–6
- Innocenti A, Zimmerman S, Ferry JG, et al. Carbonic anhydrase inhibitors. Inhibition of the zinc and cobalt gamma-class enzyme from the archaeon Methanosarcina thermophila with anions. Bioorg Med Chem Lett 2004;14:3327–31
- Alafeefy AM, Abdel-Aziz HA, Vullo D, et al. Inhibition of carbonic anhydrases from the extremophilic bacteria Sulfurihydrogenibium yellostonense (SspCA) and S. azorense (SazCA) with a new series of sulfonamides incorporating aroylhydrazone-, [1,2,4]triazolo[3,4-b][1,3,4]thiadiazinyl- or 2-(cyanophenylmethylene)-1,3,4-thiadiazol-3(2H)-yl moieties. Bioorg Med Chem 2014;22:141–7
- Del Prete S, Vullo D, Scozzafava A, et al. Cloning, characterization and anion inhibition study of the delta-class carbonic anhydrase (TweCA) from the marine diatom Thalassiosira weissflogii. Bioorg Med Chem 2014;22:531–7
- Rodrigues GC, Feijo DF, Bozza MT, et al. Design, synthesis, and evaluation of hydroxamic acid derivatives as promising agents for the management of Chagas disease. J Med Chem 2014;57:298–308
- Vullo D, Kupriyanova EV, Scozzafava A, et al. Anion inhibition study of the beta-carbonic anhydrase (CahB1) from the cyanobacterium Coleofasciculus chthonoplastes (ex-Microcoleus chthonoplastes). Bioorg Med Chem 2014;22:1667–71
- Syrjanen L, Vermelho AB, Rodrigues Ide A, et al. Cloning, characterization, and inhibition studies of a beta-carbonic anhydrase from Leishmania donovani chagasi, the protozoan parasite responsible for leishmaniasis. J Med Chem 2013;56:7372–81
- Akdemir A, Guzel-Akdemir O, Scozzafava A, et al. Inhibition of tumor-associated human carbonic anhydrase isozymes IX and XII by a new class of substituted-phenylacetamido aromatic sulfonamides. Bioorg Med Chem 2013;21:5228–32
- Guzel-Akdemir O, Akdemir A, Pan P, et al. A class of sulfonamides with strong inhibitory action against the alpha-carbonic anhydrase from Trypanosoma cruzi. J Med Chem 2013;56:5773–81
- Bilginer S, Unluer E, Gul HI, et al. Carbonic anhydrase inhibitors. Phenols incorporating 2- or 3-pyridyl-ethenylcarbonyl and tertiary amine moieties strongly inhibit Saccharomyces cerevisiae beta-carbonic anhydrase. J Enzyme Inhib Med Chem. 2013. [Epub ahead of print]. doi: 10.3109/14756366.2013.806497
- Pan P, Vermelho AB, Scozzafava A, et al. Anion inhibition studies of the alpha-carbonic anhydrase from the protozoan pathogen Trypanosoma cruzi, the causative agent of Chagas disease. Bioorg Med Chem 2013;21:4472–6
- Alafeefy AM, Isik S, Al-Jaber NA, et al. Carbonic anhydrase inhibitors. Benzenesulfonamides incorporating cyanoacrylamide moieties strongly inhibit Saccharomyces cerevisiae beta-carbonic anhydrase. Bioorg Med Chem Lett 2013;23:3570–5
- Vullo D, Leewattanapasuk W, Muhlschlegel FA, et al. Carbonic anhydrase inhibitors: inhibition of the beta-class enzyme from the pathogenic yeast Candida glabrata with sulfonamides, sulfamates and sulfamides. Bioorg Med Chem Lett 2013;23:2647–52
- Vullo D, Isik S, Del Prete S, et al. Anion inhibition studies of the alpha-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem Lett 2013;23:1636–8
- Monti SM, De Simone G, Dathan NA, et al. Kinetic and anion inhibition studies of a beta-carbonic anhydrase (FbiCA 1) from the C4 plant Flaveria bidentis. Bioorg Med Chem Lett 2013;23:1626–30
- Pan P, Vermelho AB, Capaci Rodrigues G, et al. Cloning, characterization, and sulfonamide and thiol inhibition studies of an alpha-carbonic anhydrase from Trypanosoma cruzi, the causative agent of Chagas disease. J Med Chem 2013;56:1761–71
- Akdemir A, Vullo D, De Luca V, et al. The extremo-alpha-carbonic anhydrase (CA) from Sulfurihydrogenibium azorense, the fastest CA known, is highly activated by amino acids and amines. Bioorg Med Chem Lett 2013;23:1087–90
- Nishimori I, Vullo D, Minakuchi T, et al. Restoring catalytic activity to the human carbonic anhydrase (CA) related proteins VIII, X and XI affords isoforms with high catalytic efficiency and susceptibility to anion inhibition. Bioorg Med Chem Lett 2013;23:256–60
- Vullo D, De Luca V, Scozzafava A, et al. Anion inhibition studies of the fastest carbonic anhydrase (CA) known, the extremo-CA from the bacterium Sulfurihydrogenibium azorense. Bioorg Med Chem Lett 2012;22:7142–5
- Vullo D, De Luca V, Scozzafava A, et al. The first activation study of a bacterial carbonic anhydrase (CA). The thermostable alpha-CA from Sulfurihydrogenibium yellowstonense YO3AOP1 is highly activated by amino acids and amines. Bioorg Med Chem Lett 2012;22:6324–7
- Vullo D, Luca VD, Scozzafava A, et al. The alpha-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1 is highly susceptible to inhibition by sulfonamides. Bioorg Med Chem 2013;21:1534–8
- Khalifah RG. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J Biol Chem 1971;246:2561–73
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680–5
- Guilloton MB, Korte JJ, Lamblin AF, et al. Carbonic anhydrase in Escherichia coli. A product of the cyn operon. J Biol Chem 1992;267:3731–4
- Capasso C, Supuran TC. An overview of the alpha-, beta- and gamma-carbonic anhydrases from bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria? J Enzyme Inhib Med Chem 2014. [Epub ahead of print]. doi: 10.3109/14756366.2014.910202
- Li XJ, Xie HL, Lei SJ, et al. Reduction of CAII expression in gastric cancer: correlation with invasion and metastasis. Chin J Cancer Res 2012;24:196–200
- Parkkila S, Innocenti A, Kallio H, et al. The protein tyrosine kinase inhibitors imatinib and nilotinib strongly inhibit several mammalian alpha-carbonic anhydrase isoforms. Bioorg Med Chem Lett 2009;19:4102–6

