Abstract
Arachidonic acid is an unsaturated fatty acid liberated from phospholipids of cell membranes. NSAIDs are known as targets of cyclooxygenase enzyme (COX-1, COX-2 and COX-3) in arachidonic acid metabolism. This mechanism of COX-2 in carcinogenesis causes cancer. In addition, COX-2 plays a role in the early stages of hepatocarcinogenesis. Hepatitis C virus (HCV) infection is cause of liver cirrhosis and hepatocellular carcinoma (HCC). The aim of our study was to improve effective agents against HCV. A novel series of new etodolac 1,2,4-triazoles derivatives (4a–h) have been synthesized and investigated for their activity against HCV NS5B polymerase. Compound 4a was found to be the most active with IC50 value of 14.8 µM. In accordance with these results, compound 4a was screened for anti-cancer activity on liver cancer cell lines (Huh7, Mahlavu, HepG2, FOCUS). Compound 4a showed anti-cancer activity against Huh7 human hepatoma cell line with IC50 value of 4.29 µM. Therefore, compound 4a could be considered as a new anti-cancer and anti-HCV lead compound.
Introduction
Non-steroidal anti-inflammatory drugs (NSAIDs) have recently gained attention as promising cancer chemopreventive agents. Evidence for a connection between COX-2 activation and carcinogenesis has come from a number of studiesCitation1. In addition, many epidemiological studies have also highlighted COX-2 as an important molecular target for anti-cancer therapy. Overexpression of COX-2 in tumor cell lines affect diverse mechanisms involved in carcinogenesis, such as angiogenesis, inhibition of apoptosis, stimulation of cell growth as well as the invasiveness of tumor cells. Hepatocellular carcinoma (HCC) is one of the most common cancers worldwide, accounting for approximately 1 million deaths annually and an estimated 500 000 new cases per yearCitation2. Most cases of HCC are secondary to either viral hepatitis infection (hepatitis B or C) or cirrhosisCitation3. High levels of COX-2 expression in hepatocytes may be involved in hepatocarcinogenesis following HBV and HCV infections. COX-2 upregulation has also been correlated with the presence of inflammatory cells in hepatocarcinogenesis and tumor angiogenesis in HCCCitation4. COX-2 inhibitors have displayed significant anti-proliferative effects in many human HCC cell linesCitation5. Further, investigations on animal models of liver cancer have shown that NSAIDs, including selective COX-2 inhibitors, exhibit chemopreventive as well as therapeutic effects. Etodolac, (R,S)-2-[1,8-diethyl-1,3,4-tetrahydrapyrano[3,4-b]indole-1-yl]acetic acid, is a nonsteroidal antiinflammatory drug with marked antirheumatic, analgesic and antipyretic properties. It exists as a racemic mixture of R- and S-enantiomers that are not metabolically interconvertible, of which the the S-enantiomer exhibits selective COX-2 inhibitory activityCitation6. Etodolac, at physiological doses has been reported to decrease the cell proliferative effect of β-catenin in hepatoma cellsCitation7 and also decrease PGE2 levels in HCV-associated HCCCitation8. Other studies have reported the anti-HCC properties of etodolac through mechanisms involving growth inhibition and cell cycle arrestCitation9. Thus, etodolac appears to be a promising scaffold for the development of novel anti-hepatitis C and anti-HCC agents.
Among other anti-tumor and anti-HCVagents, the 1,2,4-triazoles and pyrano[3,4-b]indoles bearing compounds have displayed promising characteristics. The five-membered heterocyclic ring of the triazoles has served as an important scaffold for many bioactive compounds, some of which have emerged as effective and non-toxic antitumor drugsCitation10,Citation11. The pyrano[3,4-b]indole derivatives, on the other hand, have been documented as potent in vitro anti-HCV NS5B polymerase inhibitorCitation12,Citation13. We recently reported on the synthesis of etodolac thiosemicarbazides and identified their anti-HCV NS5B polymerase activityCitation14.
In continuation of our efforts towards developing new anti-cancer agents with enhanced biological properties and potent anti-HCV NS5B polymerase activity, here we report on the synthesis and biological evaluation of novel etodolac 1,2,4-triazole derivatives. A total of 34 etodolac hydrazide derivatives were investigated for their activity against HCV NS5B polymerase. Selected compounds were evaluated for anti-cancer activity against a panel of 60 human tumor cell lines in addition to four hepatocellular cell lines We further report on the potential binding mode of the most active etodolac 1,2,4-triazole derivative on NS5B.
Experimental
Chemistry
General
All chemicals were purchased from Merck Co. (Darmstadt, Germany), Sigma-Aldrich (St. Louis, MO) or Fluka (Buchs, Switzerland). Reactions were monitored by thin-layer chromatography (TLC) on aluminium oxide 60F254 plates (Merck Co., Darmstadt, Germany). Melting points of the synthesized compounds were determined in Schmelzpunktgerät SMP II melting point apparatus and are presented without any corrections. The purity of the compounds was checked on TLC plates precoated with silica gel G in a solvent system comprising of petroleum ether: ethyl acetate (50:50, v/v) mixture as an eluent. The spots were located under UV light (254 nm) (t: 21°C). FT-IR spectra were recorded on Shimadzu FTIR-8400S spectrophotometer. NMR spectra were recorded on BRUKER AVANCE-DPX 400 at 400 MHz and Bruker 300 at 300 MHz (Billerica, MA) for 1H-NMR and 75 MHz for 13C-NMR (Decoupled). The chemical shifts (δ) were expressed in parts per million (ppm) downfield from tetramethylsilane (TMS) using DMSO-d6 as solvent. Data are reported as follows: chemical shift, multiplicity (b.s.: broad singlet, d: dublet; m: multiplet, s: singlet, and t: triplet), coupling constants (Hz), integration. MALDI-TOF HR-MS spectra using EI and FAB ionization techniques were performed using a Jeol JMS-700 instrument.
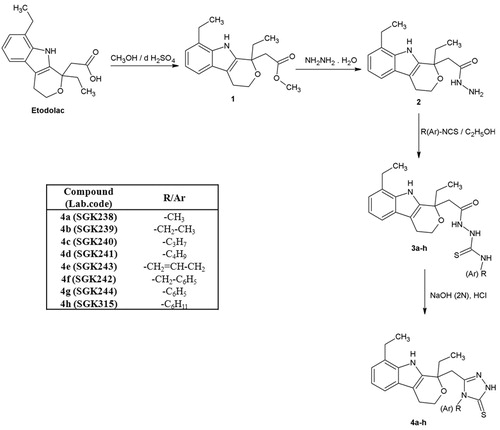
Synthesis of methyl (1,8-diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl) acetate 1, 2-(1,8-diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl)acetohydrazide 2, and 1-[2-(1,8-diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl)acetyl]-4-alkyl/aryl thiosemicarbazides (3a-h)
These etodolac derivatives were designed and prepared as described previously in our studiesCitation14–16.
General procedure for the synthesis 5-[(1,8-diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl)methyl]-4-substituted-2,4-dihydro-3H-1,2,4-triazole-3-thiones (4a–h)
A solution of 0.01 mol (3a–h) in sodium hydroxide solution (25 mL) was heated under reflux for 4 h. After cooling to room temperature, the solution was adjusted to pH 6 by concentrated hydrochloric acid. The crude product was precipitated, filtered and washed with distilled water. Pure compounds were obtained by recrystallization from ethanol.
The following compounds were prepared by an analogous procedure. The characterization of compounds 4b–e, 4g–h could be seen in Supplemental file 1.
5-[(1,8-Diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl)methyl]-4-methyl-2,4-dihydro-3H-1,2,4-triazole-3-thione (4a)
Light cream-colored solid. Yield 71%; m.p. 227–229°C; MW: 356.485; Rf × 100 value: 46.77. FT-IR (νmax, cm−1): 3279 (indole and triazole NH); 1562, 1470, 1464, 1456 (C=N, C=C); 1239 (C=S). 1H NMR (400 MHz, DMSO-d6) δ ppm: 0.64 (t, 3H, –CH2–CH3 at C1); 1.27 (t, 3H, –CH2–CH3 at C8); 1.67–2.09 (m, 2H, –CH2–CH3 at C1); 2.57–2.89 (m, 4H, –CH2 at C1 and –CH2–CH3 at C8); 3.30–3.46 (m, 5H, –CH2 at C4 and N–CH3); 3.85–3.97 (m, 2H,–CH2 at C3); 6.89–7.24 (m, 3H, Ar-H); 10.67 (s, 1H, indole NH); 13.49 (s, 1H, triazole NH). 13C NMR (100 MHz, DMSO-d6/TMS) δ ppm: 8.15 (C-12); 15.02 (C-10); 22.32 (C-4); 24.26 (C-9); 31.13 (C-11); 31.48 (N-CH3); 34.09 (C-13); 60.85 (C-3); 77.10 (C-1); 108.50 (C-5); 115.94 (C-1a); 119.26 (C-6); 120.30 (C-7); 126.32 (C-8a); 127.14 (C-8); 135.11 (C-5a); 135.63 (C-4a); 150.04 (triazol –C=N); 169.90 (C=S). HR-MS (EI+) Calculated/Found (m/z): 356.1670/356.1663 (M+) (C19H24N4OS); 324.1950/324.1943 (C19H24N4O); 281.1581/281.1633; 279.1372/279.1453; 268.1576/268.1513; 243.1623/243.1576; 229.1466/229.1409; 228.1382/228.1345 (C15H18NO); 227.1310/227.1298.
5-[(1,8-Diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl)methyl]-4-benzyl-2,4-dihydro-3H-1,2,4-triazole-3-thione (4f)
Light cream-colored solid. Yield 50%; m.p. 163–166°C; MW: 432.581; Rf×100 value: 80.60. FT-IR (νmax, cm−1): 3270 (indole and triazole NH); 1574, 1497, 1453, 1418 (C=N, C=C); 1261 (C=S). 1H NMR (400 MHz, DMSO-d6) δ ppm: 0.67 (t, 3H, –CH2–CH3 at C1); 1.26 (t, 3H, –CH2–CH3 at C8); 1.81–2.05 (m, 2H, –CH2–CH3 at C1); 2.82–3.35 (m, 6H, –CH2–CH3 at C8, –CH2– at C1 and –CH2 at C4); 3.60-3.88 (m, 2H, –CH2 at C3); 5.21–5.32 (m, 2H, N–CH2); 6.89–7.36 (m, 8H, Ar-H); 10.62 (s, 1H, indole NH); 13.67 (s, 1H, triazole NH). 13C NMR (100 MHz, DMSO-d6/TMS) δ ppm: 8.18 (C-12); 14.94 (C-10); 22.26 (C-4); 24.24 (C-9); 31.39 (C-11); 34.35 (C-13); 46.22 (N–CH2); 60.89 (C-3); 77.13 (C-1); 108.37 (C-5); 115.99 (C-1a); 119.28 (C-6); 120.31 (C-7); 126.28 (C4′); 127.11 (C2′ and C6′); 128.05 (C3′ and C5′); 129.15 (C-8 and C-8a); 135.09 (C-5a); 135.53 (C-4a); 136.32 (C1′); 149.64 (triazol –C=N); 167.46 (C=S). HR-MS (EI+) Calculated/Found (m/z): 432.1983/432.1975 (M+) (C25H28N4OS); 229.1466/229.1417; 228.1382/228.1385 (C15H18NO); 227.1310/227.1302.
Materials and methods
Anti-cancer activity
Compounds 4b and 4f were chosen as prototypes for anti-cancer evaluations at the National Institute of Health-National Cancer Institute (NIH-NCI). Growth inhibition was evaluated against a panel of 60 human tumor cell lines derived from nine neoplastic diseases (leukaemia, non-small cell lung cancer (NSCL), colon cancer, central nervous system (CNS) cancer, melanoma, ovarian cancer, renal cancer, prostate cancer and breast cancer cell lines) at 10-fold dilutionsin a primary anti-cancer assay in accordance with the protocol of the Drug Evaluation Branch, NCI, BethesdaCitation17–19.
Briefly, the tumor cell lines were seeded in 96-well microtiter plates in RPMI 1640 medium containing 5% fetal bovine serum and 2 mM L-glutamine at 37 °C, 5% CO2, 95% air and 100% relative humidity atmosphere for 24 hCitation19. Compounds were screened at a single concentration of 10−5 M for 48 h and growth inhibition was investigated by the sulforhodamine B (SRB) protein assayCitation17. The 50% inhibition was extrapolated from dose–response parameters at 10-fold dilutions of compounds as described previouslyCitation18. The percentage of growth inhibition was determined spectrophotometrically against DMSO treated controls.
HCV NS5B polymerase inhibitory activity
The effect of the compounds on HCV NS5B polymerase activity was evaluated on poly rA-U12 template- primer in the presence of [α-32P] UTP and MnCl2 as the divlent cation as described previouslyCitation20,Citation21. Reactions in the presence of the compound or DMSO were incubated at 30 °C for 1 h and terminated by the addition of 5% TCA. The nascent radiolabeled RNA was precipitated on GF-B filters and counted in a liquid scintillation counter. NS5B activity in the presence of DMSO was set at 100% and that in the presence of the compounds was determined relative to this control. Preliminary screning was conducted at 100 µM compound concetration and those exhibiting ≥50% inhibition were investigated further for their IC50 values at 8-10 concentrations of the serially diluted compounds. The IC50 values were analyzed from dose-response curves utilizing Graphpad prism 3.03 software (La Jolla, CA).
Molecular modeling
The X-ray co-crystal structure of HCV NS5B-PF868554 (PDB ID: 3FRZ) obtained from the RCSB Protein Data Bank (Piscataway, NJ) was used for docking the compounds into the NS5B thumb pocket-IICitation22. The protein structure was processed by means of default parameters mentioned in Protein Preparation Tool present in Maestro v9.0 and Impact program v5.5 (Schrödinger, Inc., New York, NY), in which the protonation states of residues were adjusted to the dominant ionic forms at pH 7.4. Refined HCV NS5B structure was further used to generate energy grid by selecting bound inhibitor (PF868554) as a reference ligand.
Docking protocol
Compounds were constructed using the fragment dictionary of Maestro 9.0 and energy minimized by Macromodel program v9.7 (Schrödinger, Inc., New York, NY). The energy minimization involved steepest descent followed by truncated Newton conjugate gradient application of OPLSAA force field parameters (convergence criteria of 0.001 rmsd was used). The low-energy 3D structures of etodolac analogs were generated with the help of LigPrep v2.3 (Schrödinger Inc., New York, NY): different protonation states at physiological pH, all possible tautomers, ring conformations and stereoisomersCitation23. The resulting LigPrep derived structures were used for docking simulations. The X-ray co-crystal structure of HCV NS5B-PF868554 (PDB ID: 3FRZ) was used for docking into thumb pocket-IICitation22. Protein refinement without crystallographic water molecules, energy grid generation using bound ligand as a reference, and “Extra Precision” (XP) Glide docking v5.0 (Schrödinger, Inc., New York, NY) was performed with the default parameters. The top scoring pose of compound 4a within the TP-II was used for graphical analysis. All computations were carried out on a Dell Precision 470 n dual processor with the Linux OS (Red Hat Enterprise WS 4.0, Raleigh, NC).
NCI-Sulforhodamine B (SRB) anti-cancer assay of etodolac triazole, SGK-238 on human liver cancer cells
Human liver cancer cells (Huh7, Mahlavu, HepG2, FOCUS) were cultured in 96-well plates (1000–3000 cell/well) and grown for 24 h. Then the cells were treated with increasing concentrations of the compound (2.5–40 μM). After 72 h treatment, the growth medium was aspirated and cells were washed with 1×PBS (CaCl2-, MgCl2-free) (Gibco, Invitrogen, Grand Island, NY) followed by the addition of 50 μl cold 10% (v/v) trichloroacetic acid (TCA) and incubated 1 h at 4°C. TCA solution was then aspirated and the micro-plates were washed five times with deionized water and left air-dry. Finally, 50 μl of 0.4% (m/v) sulforhodamine (Sigma-Aldrich) in 1% acetic acid solution was added to each well and the plates were incubated for 10 min at room temperature. In order to remove the unbound stain, the plates were washed five times with 1% acetic acid and left to air-dry. The bound sulforhodamine B was then solubilized using 10 mM Tris-base. The absorbance was measured at 515 nm.
Results and discussion
Chemistry
(R,S)-2-[1,8-Diethyl-1,3,4-tetrahydrapyrano[3,4-b]indole-1-yl]acetic acid (etodolac) was chosen as the starting compound to design 1,2,4-triazole-3-thiones derivatives. Synthesis of triazole derivatives (4a–h) required stepwise reactions starting with etodolac hydrazide. The synthetic route for the title compounds is shown in . In our previous study, we synthesized methyl 2-(1,8-diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl) acetate (1) and 2-(1,8-diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl)acetohydrazide (2)Citation15. The reaction of corresponding hydrazide 2 with respective alkyl/aryl isothiocyanates yielded etodolac thiosemicarbazides (3a–h)Citation14,Citation16. In present study, smooth cyclization of etodolac thiosemicarbazides under alkaline conditions afforded the 5-[(1,8-diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl)methyl]-4-substituted-2,4-dihydro-3H-1,2,4-triazole-3-thiones (4a–h) in moderate to good yieldsCitation16. These compounds are original. All the newly synthesized etodolac 1,2,4-triazoles are air-stable solids and soluble in DMSO at ambient temperature. The purity of the synthesized compounds was checked by thin layer chromatography (TLC) and microanalysis. The structures of obtained compounds were determined on the basis of FT-IR, 1H-NMR, 13C-NMR and HR-MS spectral data and all of the compounds have satisfactory analyses for their proposed structures. 1H-NMR soctral results for etodolac triazoles together with hydrogen assignments. 13C-NMR and HR-MS spectra results are also demonstrated in Experimental Section.
The refluxing of etodolac thiosemicarbazides in aqueous NaOH transformed into the target compounds (4a–h) was indicated in the IR spectra by appearance of a single absorption for NH in the region 3160–3349 cm−1 and disapperance of a comparatively strong band in the region 1674–1688 cm−1 for the carbonyl of hydrazides, indicated the formation of triazoles. The absorption bands at 1576–1416 cm−1 are due to the presence of –C=N– stretch of the triazole ring system. Furthermore, the appearance of a C=S absorption band in the region 1215–1262 cm−1 indicated that the triazoles are in their thione form. The existence of triazoles in thione form was also indicated by the absence of SH stretching in the characteristic region of 2550–2600 cm−1Citation24.
1H-NMR data was also in agreement with the proposed structures of all newly synthesized etodolac triazoles. Moreover, in the 1H-NMR spectra of etodolac triazoles, additional signals due to the NH group were appeared in the region of δ 13.38–13.82 ppm, whereas S–H proton at 1.0–3.7 ppm was not detected which also confirmed the formation of their thionic form Citation25. Triazole NH resonances were observed to be shifted to downfield because of strong intermolecular hydrogen bonding as previously reportedCitation26,Citation27. 1H-NMR spectra of the etodolac thiosemicarbezides (3a–h) showed three proton signals typical for linked to N1–N2 and N4 nitrogens in the 7.03–10.55 ppm range in our previous study, whereas in the 1H-NMR spectra of etodolac triazoles (4a–h) the singlet peak due to the proton of NH group appeared, which confirmed the succesful formation of the desired triazoles. All other aromatic and aliphatic protons for obtained compounds were observed at expected chemical shifts and integral values.
13C-NMR spectra of compounds exhibited additional signals due to triazole moiety at the expected chemical shift values. The carbon of triazole, C=S group and C=N group had a typical signals at about 165.49–170.05 and 149.33–150.04 ppm, respectively. The peaks resonated at region of 165.49–170.05 ppm in the 13C-NMR spectra of these compounds, assigned for C=S, confirming thione form of triazoleCitation28. The 13C-NMR spectra of the compounds displayed the appropriate number of resonances that exactly assembled the number of carbon atoms. The etodolac derivatives may occur in thione or thiol form. However, in our case the signal of C=S group in the 13C-NMR spectra and disappearance of S–H proton in the 1H-NMR spectra of etodolac triazoles (4a–h) indicated that they were existed in a solution predominantly in thione form. The thione-thiol tautomerism was not observed for etodolac triazoles.
High-resolution mass spectra (HR-MS) confirmed the molecular weights and empirical formulae of the compounds and fragments, with less than 5 mmu bias between the calculated and experimental m/z values of either the molecular or the fragment ions. The ionization mode was electron impact (EI) for all triazole derivatives. Etodolac triazoles gave relatively stable molecular ion peaks in the corresponding mass spectra. In addition, the fragmentation pattern for all compounds was supported in the mass spectra. The fragment ion peaks especially [M − N2]+, [M − HCNS]+ , [M − RNCS]+ and [M − S] + are the diagnostic peaks for 1,2,4-triazoles ringsCitation24,Citation29. IR, 1H-NMR, 13C-NMR and mass spectral data are in agreement with the proposed structures of all newly synthesized compounds.
Biological evaluation
Anti-cancer activity
The growth inhibition properties of the compounds selected by NCI, USA, was performed in accordance with the protocol of their Drug Evaluation Branch Selection, and is available online at the DTP web site (http://www.dtp.nci.nih.gov/docs/misc/common_files/guidelines.html)Citation18,Citation30. The cytotoxic and/or growth inhibitory effects of the compounds with ethyl and benzyl substituent, 4b and 4f chosen as prototypes were tested at a concentration of 10−5 M against the full panel of 60 human tumour cell lines derived from nine neoplastic diseases. The percentage of growth was determined spectrophotometrically and compared against DMSO-treated controls. The one-dose assay facilitates detection of both growth inhibition (values between 0 and 100) and lethality (values less than 0). It means that a value of 100 means no growth inhibition. A value of 40 would mean 60% growth inhibition. A value of 0 means no net growth over the course of the experiment. A value of -40 would mean 40% lethality. A value of −100 means all cells are dead. Compounds with cell lines appearing on positive side of mean graph display high growth inhibition of the specific cancer cells. The one-dose data will be described as a mean graph of the percent growth of treated cells. Mean graphs were established for each effect, with bars showing the deviation of individual tumour cell lines from the over all mean value for all the tested cells. In the mean graph, the centre point is the mean of the growth inhibition (GI) percentage over all cell lines. Bars that point to the right are cell lines where the inhibition is greater than the average, while bars that point to the left are cell lines where the inhibition is less than the average. Compounds with cell lines appearing on positive side of mean graph display high growth inhibition of cancer cells of that particular cancer. In , the results of in vitro anti-cancer screening of 4b and 4f are summarized. Tested compounds had moderate growth inhibition effects, which means they had not pass the 5-dose assay. They were unable to reduce the growth of any of the cell lines to 32% or less.
Table 1. The anticancer screening data of etodolac triazoles, 4b and 4f.
5-[(1,8-Diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl)methyl]-4-benzyl-2,4-dihydro-3H-1,2,4-triazole-3-thione, 4f demonstrated 46.15, 45.39, 46.49, 50.03% growth percent at 10−5 M on CNS cancer SNB-75, melanoma UACC-257, renal cancer CAKI-1, prostate cancer PC-3 cancer cell lines, respectively. Moreover, compound 4b had a strong cytotoxicity against melanoma MALME-3 M and compound 4f had a strong cytotoxicity against melanoma MALME-3 M and leukemia HL-60(TB) cell lines. There are also several reports in the literature on etodolac which were observed to inhibit melanoma by cyclooxygenase inhibitorsCitation31. Our findings indicated that 1,2,4-triazoles derivatived from etodolac keep going on effect of melanoma.
Anti-HCV NS5B polymerase activity
Previously synthesized compoundsCitation15, of etodolac ester (Lab. Code: SGK-196), hydrazide (Lab. Code: SGK-197) etodolac hydrazones (Lab. Code: SGK 198-210), 4-thiazolidinones (Lab. Code: SGK 212-214, SGK 216-221, SGK223) and newly synthesizd compounds of etodolac triazoles (4a–h,) were evaluated for their inhibition of hepatitis C virus NS5B RNA dependent RNA polymerase activityCitation32. The anti-NS5B polymerase activity of the etodolac 1,2,4-triazoles () was investigated in vitro against recombinant NS5BCΔ21 1b by a primer-dependent elongation assay as described previouslyCitation20,Citation21. All compounds were subjected to preliminary screening at 100 μM concentration and candidates displaying ≥50% inhibition of NS5B polymerase activity at this concentration were further investigated for their IC50 value. Etodolac, the parent molecule, included in this investigation for comparison, yielded ∼10% inhibition of NS5B polymerase, while its derivatives displayed ∼5.0–80.0% inhibition of NS5B polymerase activity at this concentration. Of these, all 11 etodolac 4-thiazolidinones proved to be weak NS5B polymerase inhibitors with ≤50% anti-NS5B activity, while two etodolac hydrazide-hydrazones and five etodolac triazoles appeared more promising displaying ≥50% inhibition of NS5B polymerase activity. The triazoles were overall more active than the hydrazide-hydrazones, displaying IC50 values between 14.0 and 26.0 µM, with the exception of compound 4e (IC50= 40.0 µM). Among these, compound 4a (SGK 238) with IC50 value of 14.8 μM was the most active of the etodolac triazoles. General structure and substituents of etodolac hydrazide hydrazones and etodolac 4-thiazolidinones which were synthesized in our previous studyCitation15 are given in Supplemental file 2.
Table 2. Anti-HCV NS5B RdRp activity of etodolac derivatives.
To evaluate the probable binding conformation of the most potent chiral etodolac derivatives, we performed docking study using Glide docking software. Since the structurally related pyranoindoles have been previously shown to inhibit NS5B activity through binding to TP-II, we performed docking calculations at TP-II siteCitation13,Citation22. Analysis of the binding energy data for the docked conformations of R- versus S-isomers of the most active compound of 5-[(1,8-diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl)methyl]-4-methyl-2,4-dihydro-3H-1,2,4-triazole-3-thione, 4a (Glidescore for (R) = −2.94 kcal/mol and for (S) = −4.58 kcal/mol), showed that S-isomer bind at least by 1 kcal/mol better than their R-counterpart. The binding mode of (S)-isomer of the etodolac derivative 4a within the TP-II of HCV NS5B polymerase is shown in (please view this figure in color mode in the online version). The spCitation2 hybridized nitrogen atom of the triazolethione ring forms a water (HOH677) mediated hydrogen bond with the backbone –NH of Tyr477 (=N⋯H2O⋯HN–Tyr477). The pyran ring oxygen atom also involved in water (HOH677)-mediated hydrogen bonding interaction with the backbone –NH of Tyr477 (O⋯H2O677⋯HN–Tyr477). This pyran ring oxygen atom is also found at electrostatic interacting distance from the backbone –NH of Ser476 (–O⋯HN-Ser476, 3.58 Å). The N-methyl, the thioxo and the –NH groups of the triazolethione ring are solvent-exposed. The ethyl substituent on indole nucleus forms hydrophobic interactions with the side chains of Leu419, Met423 and Leu497. The indole nucleus is stabilized by hydrophobic interactions with the side chains of Met423, Tyr477, Leu497 and Trp528. The ethylpyran moiety is mainly stabilized by hydrophobic contacts with the side chain of Tyr477.
Figure 2. Glide-XP predicted binding model of compound (S)-4a within the TP-II of HCV NS5B polymerse. Amino acid residues are shown as stick model with the atoms colored as carbon – green, hydrogen – white, nitrogen – blue oxygen – red and sulfur – yellow whereas inhibitor is shown as ball and stick model with the same color scheme as above except carbon atoms are represented in orange. Water molecules are shown as stick model. Dotted black line indicates hydrogen bonding interaction whereas dotted pink line indicates potential electrostatic contact with distances in Å (please view this figure in color mode in the online version).
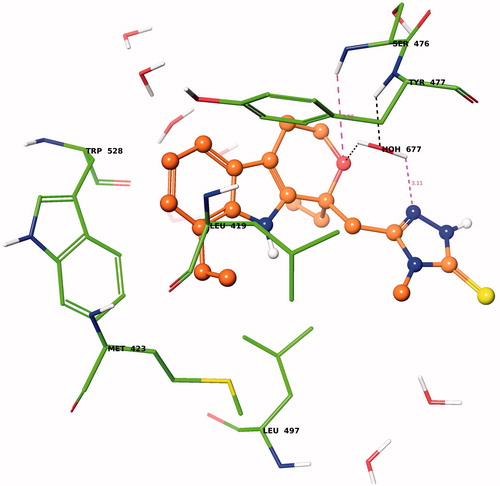
The binding mode of (R)-4a was found to be completely different and showed no key hydrogen bonding interactions with the residues of TP-II such as Ser476 and Tyr477. Therefore binding energy data and predicted binding conformation together suggest that NS5B inhibitory activity of racemic 4a could be due to S-isomer. Future study will focus on separation of each enantiomer as well as optimization of compound 4a.
Anti-cancer activity of the etodolac triazole, 4a against human liver cancer cells
Compound 4a, which exhibited the highest inhibitory activity against HCV NS5B polymerase, was evaluated for its ability to inhibit the growth of liver cancer employing four prototype liver cancer cell lines, Huh7, Mahlavu, HepG2 and FOCUS. Towards this goal, we carried out an anti-cancer drug-screening method based on a sulforhodamine B assay (SRB) in triplicate to determine their IC50 valuesCitation17.
The cells were treated with increased concentration from 2.5 to 40 μM of the compound 4a and incubated for 72 h. Absorbance values were obtained and normalized to DMSO control. The result of anti-cancer analysis of this compound is summarized in . However, the IC50 values were in micromolar concentrations with 4a against each human liver cancer cells (). 5-[(1,8-Diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl)methyl]-4-methyl-2,4-dihydro-3H-1,2,4-triazole-3-thione, 4a displayed the best anti-cancer activity, with IC50 value of 4.29 against Huh7 cell line. Considering the anti-cancer activity of this etodolac triazole on the hepatoma cell lines, we further analyzed the cellular activity as a promising candidate anti-cancer agent.
Figure 3. Percent growth inhibition graphs of compound 4a, SGK-238 on liver cancer cell lines (Huh7, Mahlavu, HepG2, FOCUS).
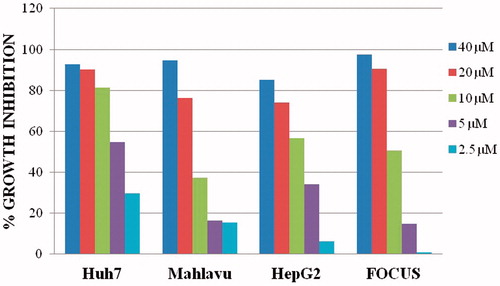
Table 3. IC50 values in µM concentrations for compound 4a with 72 h of treatment on liver cancer cell linesa.
Conclusions
In the current study, a series of novel etodolac 1,2,4-triazoles were synthesized and evaluated for their anti-cancer activity and inhibition of hepatitis C virus NS5B RNA dependent RNA polymerase activity. After a preliminary survey, 5-[(1,8-diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl)methyl]-4-ethyl-2,4-dihydro-3H-1,2,4-triazole-3-thione, 4b and 5-[(1,8-diethyl-1,3,4,9-tetrahydropyrano[3,4-b]indole-1-yl)methyl]-4-benzyl-2,4-dihydro-3H-1,2,4-triazole-3-thione, 4f were selected by the NCI for anti-cancer screening. These etodolac triazoles were found to show significant inhibition on CNS cancer SNB-75, melanoma UACC-257, renal cancer CAKI-1, prostate cancer PC-3 cancer cell lines. In this present study, we have also evaluated inhibition of hepatitis C virus NS5B RNA dependent RNA polymerase activity of etodolac derivatives. Among these, the most active triazole, compound 4a (SGK 238), with methyl substituent displayed IC50 values of 14.8 μM. Notably, this study reports for the first time the anti-HCV NS5B activity of etodolac 1,2,4-triazoles. Molecular docking and binding mode investigations suggest that the 1,2,4-triazole scaffold may be optimized for generating more effective analogues with improved anti-NS5B potency. In order to analyze anti-cancer property of the active compound 4a, anti-cancer assay was investigated on diverse human cancer cell lines. Based on these studies, the IC50 value of compound 4a was analyzed on hepatoma cells. We are now in the process of synthesizing modified analogues of the lead compound in order to generate more effective hepatitis C virus NS5B polymerase inhibitors and candidate anti-cancer agents.
Supplementary material available online.
Supplemental file 1 and 2.
Supplemental Material.pdf
Download PDF (118 KB)Acknowledgements
We thank the Division of Cancer Research, National Cancer Institute, Bethesda, MD, for the anti-cancer activity screening. The authors are grateful to Dr. Jürgen Gross from the Institute of Organic Chemistry, University of Heidelberg, for his generous help on obtaining HR-EI/FAB mass spectra of the synthesized compounds. Etodolac was supplied by Bilim Pharmaceutical Industry Inc.
Declaration of interest
The authors declare no conflicts of interest. The authors alone are responsible for the content and writing of this article. This work was supported by The Scientific and Technical Research Council of Turkey (TÜBİTAK), Research Fund Project Number: SBAG-HYD-339 (108S257) to S.G.K. HCV NS5B inhibtion studies were partly supported by the National Institute of Health Research Grants DK066837 and CA153147 to N.K-B.
Notes
* This work was partly presented at the 2nd International BAU-Drug Design Congress, Nowel Methods and Emerging Targets in Drug Design&Patented Drug Development, Istanbul-TURKEY, 17-19 April 2014.
References
- Hawk ET, Viner JL, Dannenberg A, DuBois RN. COX-2 in cancer – a player that's defining the rules. J Natl Cancer Inst 2002;94:545–6
- Montalto G, Cervello M, Giannitrapani L, et al. Epidemiology, risk factors, and natural history of hepatocellular carcinoma. Ann N Y Acad Sci 2002;963:13–20
- Carr BI. Hepatocellular carcinoma: current management and future trends. Gastroenterology 2004;127:218–24
- Koga H, Sakisaka S, Ohishi M, et al. Expression of cyclooxygenase-2 in human hepatocellular carcinoma: relevance to tumor dedifferentiation. Hepatology 1999;29:688–96
- Küçükgüzel ŞG, Coşkun İ, Aydın S, et al. Synthesis and characterization of celecoxib derivatives as possible anti-inflammatory, analgesic, antioxidant, anticancer and anti-HCV agents. Molecules 2013;18:3595–614
- Demerson CA, Humber LG, Abraham NA, et al. Resolution of etodolac and antiinflammatory and prostaglandin synthetase inhibiting properties of the enantiomers. J Med Chem 1983;26:1778–80
- Behari J, Zeng G, Otruba W, et al. R-Etodolac decreases beta-catenin levels along with survival and proliferation of hepatoma cells. J Hepatol 2007;46:849–57
- Liu W, Nakamura H, Tsujimura T, et al. Chemoprevention of spontaneous development of hepatocellular carcinomas in fatty liver Shionogi mice by a cyclooxygenase-2 inhibitor. Cancer Sci 2006;97:768–73
- Cheng J, Imanishi H, Liu W, et al. Involvement of cell cycle regulatory proteins and MAP kinase signaling pathway in growth inhibition and cell cycle arrest by a selective cyclooxygenase 2 inhibitor, etodolac, in human hepatocellular carcinoma cell lines. Cancer Sci 2004;95:666–73
- Duran A, Doğan HN, Rollas S. Synthesis and preliminary anticancer activity of new 1,4-dihydro–3-(3-hydroxy-2-naphthyl)-4-substituted-5H-1,2,4-triazoline-5-thiones. Il Farmaco 2002;57:559–64
- Li Z, Gu Z, Yin K, Zhang R, et al. Synthesis of substituted-phenyl-1,2,4-triazol-3-thione analogues with modified D-glucopyranosyl residues and their antiproliferative activities. Eur J Med Chem 2009;44:4716–20
- Gopalsamy A, Lim K, Ciszewski G, et al. Discovery of pyrano[3,4-b]indoles as potent and selective HCV NS5B polymerase inhibitors. J Med Chem 2004;47:6603v8
- LaPorte MG, Draper TL, Miller LE, et al. The discovery and structure-activity relationships of pyrano[3,4-b]indole based inhibitors of hepatitis C virus NS5B polymerase. Bioorg Med Chem Lett 2010;20:2968–73
- Çıkla P, Arora P, Basu A, et al. etodolac thiosemicarbazides: a novel class of hepatitis C virus NS5B polymerase inhibitors. Marmara Pharm J 2013;17:138–46
- Çıkla P, Özsavcı D, Bingöl-Özakpınar Ö, et al. Synthesis, cytotoxicity, and pro-apoptosis activity of etodolac hydrazide derivatives as anticancer agents. Arch Pharm (Weinheim) 2013;346:367–79
- Küçükgüzel ŞG, Süzgün P. Synthesis of thiosemicarbazides and triazoles derived from etodolac. Patent WO 2014/003694 A1; 2014
- Skehan P, Storeng R, Scudiero D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst 1990;82:1107–12
- Grever MR, Schepartz SA, Chabner BA. The National Cancer Institute: cancer drug discovery and development program. Semin Oncol 1992;19:622–38
- Weinstein JN, Myers TG, O’Connor PM, et al. An information-intensive approach to the molecular pharmacology of cancer. Science 1997;275:343–9
- Kaushik–Basu N, Bopda-Waffo A, Basu A, et al. 4-Thiazolidinones: a novel class as hepatitis C virus NS5B polymerase inhibitors. Front Biosci 2008;13:3857–68
- Kaushik-Basu N, Bopda-Waffo A, Talele TT, et al. Identification and characterization of coumestans as novel HCV NS5B polymerase inhibitors. Nucleic Acids Res 2008;36:1482–96
- Li H, Tatlock J, Linton A, et al. Discovery of (R)-6-cyclopentyl-6-(2-(2,6-diethylpyridin-4-yl)ethyl)-3-((5,7-dimethyl-[1,2,4]triazolo [1,5-a]pyrimidin-2-yl) methyl)-4-hydroxy-5,6-dihydropyran-2-ne (PF-00868554) as a potent and orally available hepatitis C virus polymerase inhibitor. J Med Chem 2009;52:1255–8
- Nichols DB, Leão RA, Basu A, et al. Evaluation of coumarin and neoflavone derivatives as HCV NS5B polymerase inhibitors. Chem Biol Drug Des 2013;81:607–14
- Küçükgüzel ŞG, Rollas S, Erdeniz H, Kiraz M. Synthesis, characterization and antimicrobial evaluation of ethyl 2-arylhydrazono-3-oxobutyrates. Eur J Med Chem 1999;34:153–60
- Rollas S, Kalyoncuoğlu N, Sur-Altiner D, Yeğenoğlu Y. 5-(4-Aminophenyl)-4-substituted-2,4-dihydro-3H-1,2,4-triazole-3-thiones: synthesis and antibacterial and antifungal activities. Pharmazie 1993;48:308–9
- Küçükgüzel İ, Küçükgüzel ŞG, Rollas S, et al. Synthesis of some 3-(arylalkylthio)-4-alkyl/aryl-5-(4-aminophenyl)-4H-1,2,4-triazole derivatives and their anticonvulsant activity. Il Farmaco 2004;59:893–901
- Küçükgüzel ŞG, Küçükgüzel İ, Tatar E, et al. Synthesis of some novel heterocyclic compounds derived from diflunisal hydrazide as potential anti-infective and anti-inflammatory agents. Eur J Med Chem 2007;42:893–901
- Salgın-Gökşen U, Gökhan-Kelekçi N, Göktaş Ö, et al. 1-Acylthiosemicarbazides, 1,2,4-triazole-5(4H)-thiones, 1,3,4-thiadiazoles and hydrazones containing 5-methyl-2-benzoxazolinones: synthesis, analgesic-anti-inflammatory and antimicrobial activities. Bioorg Med Chem 2007;15:5738–51
- Küçükgüzel I, Tatar E, Küçükgüzel SG, et al. Synthesis of some novel thiourea derivatives obtained from 5-[(4-aminophenoxy)methyl]-4-alkyl/aryl-2,4-dihydro-3H-1,2,4-triazole-3-thiones and evaluation as antiviral/anti-HIV and anti-tuberculosis agents. Eur J Med Chem 2008;43:381–92
- Alley MC, Scudiero DA, Monks A, et al. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res 1988;48:589–601
- Kast RE. Melanoma inhibition by cyclooxygenase inhibitors: role of interleukin-6 suppression, a putative mechanism of action, and clinical implications. Med Oncol 2007;24:1–6
- Küçükgüzel ŞG, Süzgün P, Arora P, et al. Etodolac derivatives as HCV NS5B polymerase inhibitors. Patent WO 2014/003693 A1; 2014