Abstract
Carbonic anhydrases (CAs, EC 4.2.1.1) catalyze the CO2 hydration/dehydration reversible reaction: CO2 + H2O ⇄ + H+. Living organisms encode for at least six distinct genetic families of such catalyst, the α-, β-, γ-, δ-, ζ- and η-CAs. The main function of the CAs is to quickly process the CO2 derived by metabolic processes in order to regulate acid-base homeostasis, connected to the production of protons (H+) and bicarbonate. Few data are available in the literature on Antarctic CAs and most of the scientific information regards CAs isolated from mammals or prokaryotes (as well as other mesophilic sources). It is of great interest to study the biochemical behavior of such catalysts identified in organism living in the Antarctic sea where temperatures average −1.9 °C all year round. The enzymes isolated from Antarctic organisms represent a useful tool to study the relations among structure, stability and function of proteins in organisms adapted to living at constantly low temperatures. In the present paper, we report in detail the cloning, purification, and physico-chemical properties of NcoCA, a γ-CA isolated from the Antarctic cyanobacterium Nostoc commune. This enzyme showed a higher catalytic efficiency at lower temperatures compared to mesophilic counterparts belonging to α-, β-, γ-classes, as well as a limited stability at moderate temperatures.
Introduction
Carbon dioxide (CO2) is an important molecule in all life processes and is generated in high amounts in all organisms, which developed catalysts for its transformation into protons and bicarbonatesCitation1–11. These catalysts are the enzymes known as carbonic anhydrases (CAs, EC 4.2.1.1) which catalyze the CO2 hydration/dehydration reversible reaction: CO2 + H2O ⇄ + H+Citation1,Citation3–6,Citation8,Citation12–19. Living organisms encode for at least six distinct genetic families of such catalyst, the α-, β-, γ-, δ-, ζ- and η-CAsCitation3,Citation10,Citation15,Citation20–25. The main function of the CAs is to quickly process the CO2 derived by metabolic processes in order to regulate acid–base homeostasis, connected to the production of protons (H+) and bicarbonate, also intervening in the secretion of electrolytes and biosynthetic processes in many cell typesCitation1–11. In fact, pH regulation is a tightly controlled process in most biochemical reactions due to the strong pH dependence of enzyme activityCitation4,Citation26–47. Mesophilic CAs are considered among the most-active enzymes known, with a catalytic efficiency approaching the limit of diffusion-controlled processesCitation24,Citation48–76.
It is of great interest to study the biochemical behavior of such catalysts identified in organism living in the Antarctic sea where temperatures average −1.9 °C all year roundCitation77–84. Antarctic organisms have evolved several strategies to overcome the effect of low temperatures on catalytic efficiency. They can follow different strategies to achieve cold adaptation at the metabolic level, such asCitation77,Citation78,Citation80,Citation81,Citation83,Citation84: (a) the increase of enzyme production in order to compensate for reduced kinetic efficiency; (b) the expression of enzymes with relatively higher substrate turnover and the capacity to maintain ligand-binding properties at low temperatures; and (c) cold-adapted enzymes evolved higher Km values (at any measured temperature) compared to their warm-adapted orthologous. Most of these strategies involve alteration of various specific features of the enzyme, in particular functionality and stabilityCitation77,Citation78,Citation80,Citation81,Citation85–88. They also represent the major hallmarks of psychrophily and have been shown to be specific from organism to organism. Probably, cold adaptation traits do not follow specific set patterns and perhaps are currently in a state of continuous evolution influenced by the environmental conditions, such as low temperature, high salinity.
Few data are available in the literature on Antarctic CAs and most of the scientific information regards CAs isolated from mammals or prokaryotes (as well as other mesophilic sources), which are active at physiological temperatures of around 37 °C. Recently, our group studied CAs from microorganisms living in extreme habitats, such as thermophiles living at temperatures ranging from 70 to 110 °CCitation1,Citation9,Citation10,Citation25,Citation89–95. These enzymes were highly active for the hydration of CO2 to bicarbonate and protons and possessed an inhibition profile (with anions and sulfonamides) very similar to those of the mesophilic human (h) cytosolic isoform hCA II. In this context, we started to study CAs from psychrophilesCitation96,Citation97. Antarctica can be considered as a huge natural laboratory and many scientists share the common interest of elucidating the evolutionary mechanisms developed by Antarctic organisms in order to adapt to this very extreme habitatCitation77,Citation78. The enzymes isolated from Antarctic organisms represent a useful tool to study the relations among structure, stability and function of proteins in organisms adapted to living at constantly low temperaturesCitation77,Citation78. Recently, we studied the catalytic activity of a γ-CA, named NcoCA, identified in the genome of the diazotrophic cyanobacteria belonging to the genus Nostoc, isolated from the freshwaters of Antarctic lakes, which form colonies composed of filaments of moniliform cells in a gelatinous sheathCitation96,Citation97. The enzyme showed a significant catalytic activity for the physiologic reaction, CO2 hydration to bicarbonate and protons, with a kcat of 9.5 × 105 s−1 and a kcat/Km of 8.3 × 107 M−1 s−1, being the most catalytically efficient γ-CA investigated so far. Our groups also carried out an extensive study using inorganic/organic anions and aromatic/heterocyclic sulfonamides inhibitors of NcoCA. The best NcoCA inhibitors detected so far were diethyldithiocarbamate (KI of 0.80 mM) as well as sulfamide, sulfamate, phenylboronic acid and phenylarsonic acid (KI in the range of 70–90 mM); while inhibitors belonging to sulfonylated sulfanilamide derivatives possessing elongated molecules, aminobenzolamide, acetazolamide, benzolamide, dorzolamide, brinzolamide and topiramate, showed inhibition constants in the range of 40.3–92.3 nM. In the present paper, we report in detail the cloning, purification, and physico-chemical properties of NcoCA.
Materials and methods
Gene identification
The identification of Nostoc γ-CA (NcoCA) was performed at the link http://www.ncbi.nlm.nih.gov/genomeCitation98 using in the keyword fields the word: “Nostoc”. The γ-CA of Nostoc sp. was identified running the “BLAST” programCitation98 using as query sequences CAM or CAMH (γ-CAs from Methanosarcina thermophila)Citation99,Citation100.
Construct preparation, protein expression and purification
Our groups designed the synthetic NcoCA DNA sequence and included the restriction enzyme NdeI and XhoI at the 5′ and 3′ end of the NcoCA gene, respectively. The gene was optionally optimized using the Gene Optimizer (Life-Technologies, Carlsbad, CA) software for maximum protein production. The GeneArt Company, specialized in gene synthesis, produced the aforementioned synthetic gene, which was provided in a standard vector (sub-cloning vector). The plasmid was amplified into E. coli DH5 α cells. The NcoCA DNA fragments were separated on 1% agarose gel. The recovered NcoCA gene and the linearized expression vector (pET15-b) were ligated by T4 DNA ligase to form the expression vector pET15-b/NcoCA. At the N-terminal sequence of NcoCA, pET15-b/NcoCA construct expressed a short polypeptide containing six histidines to facilitate the purification of the target protein. In order to confirm the integrity of the NcoCA gene and the fact that no errors occurred at the ligation sites, the vector containing the fragment was sequenced. Arctic Express competent cells (Agilent Technologies, La Jolla, CA) were transformed with pET15-b/NcoCA expression vector. Several transformant colonies were picked and grown overnight in LB containing 20 mg/ml of gentamicin and 100 mg/ml of ampicillin for selecting the expression plasmid at 37 °C. Then, cells were grown without antibiotic selection at 30 °C and induced at an A600 of 0.6–1 with 1 mM isopropylthiogalactopyranoside (IPTG). Zn(SO4) was added after 30 min. After additional growth for 16 h at 20 °C, cells were harvested, resuspended in 10 mM buffer Tris/HCl, pH 8.3 and disrupted by sonication at 4 °C. Following centrifugation, soluble protein extract was loaded onto a His-select HF nickel affinity gel column (GE Healthcare, Little Chalfont, Buckinghamshire, UK), then washed with five column volumes with His binding buffer (20 mM NaH2PO4, 500 mM NaCl, 20 mM Imidazole, pH 8). The His-tagged protein was eluted from the column with elution buffer (20 mM NaH2PO4, 500 mM NaCl, 250 mM Imidazole, pH 8). The purified recombinant NcoCA was dialyzed over-night against 10 mM buffer Tris/HCl, pH 8.3. At this stage of purification the enzyme was at least 90% pure and the obtained recovery was of 2 mg of the recombinant Antarctic γ-CA.
Sequence analysis
Multialignment of amino acid sequences was performed using the program MUSCLE (MUltiple Sequence Comparison by Log- Expectation), a new computer program for creating multiple alignments of protein sequenceCitation101.
SDS-PAGE
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed as described by Laemmli using 12% gelsCitation102.
Protonography
Briefly, wells of 12% SDS-gel were loaded with bCA, NcoCA and PgiCA mixed with loading buffer without 2-mercaptoethanol and without boiling the samples, in order to avoid protein denaturation. The gel was run at 180 V until the dye front ran off the gel. Following the electrophoresis, the 12% SDS-gel was subject to protonography to detect the PgiCA and NcoCA hydratase activity on the gel as described by Capasso and coworkersCitation22,Citation103,Citation104.
Assay for carbonic anhydrase with CO2 as substrate
Colorimetric method
CA activity assay was a modification of the procedure described by Chirica et al.Citation105. The assay was based on monitoring the pH variation due to the catalyzed conversion of CO2 to bicarbonate. Bromothymol blue was used as the indicator of pH variation. The assay was performed at 0 °C adding 1.0 mL ice-cold CO2-saturated water to 1.0 mL mixtures of 25 mM Tris-SO4 buffer containing different amounts of the enzyme. The CO2-satured solution was prepared by bubbling CO2 into 100 mL of distilled water for approximately 3 h. The CO2 solution was chilled in an ice-water bath. To test the activity of the enzyme, 1 mL of 25 mM Tris, pH 8.3, containing bromothymol blue as a dye (to give a distinct and visible blue color) was added to two test tubes chilled in an ice bath. A volume of 10 to 50 μL of the enzyme solution (e.g. cell extract or purified enzyme) was added to one tube, and an equivalent amount of buffer was added to the second tube as control. One milliliter of CO2 solution was added very quickly and simultaneously a stopwatch was started. The time required for the solution to change from blue to yellow was recorded (transition point of bromothymol blue is pH 6–7.6). The production of hydrogen ions during the CO2 hydration reaction lowers the pH of the solution until the color transition point of the dye is reached. The time required for the color change is inversely related to the quantity of carbonic anhydrase present in the sample. Detecting the color change is somewhat subjective, but the error for triple measurements was in the range of 0–1 s difference for the catalyzed reaction. Wilbur–Anderson units were calculated according to the following definition: One Wilbur–Anderson unit (WAU) of activity is defined as (T0 − T)/ T, where T0 (uncatalyzed reaction) and T (catalyzed reaction) are recorded as the time (in seconds) required for the pH to drop from 8.3 to the transition point of the dye in a control buffer and in the presence of enzyme, respectively.
Sopped-flow technique
An applied photophysics stopped-flow instrument was used for assaying the CA catalyzed CO2 hydration activityCitation106. Phenol red (at 0.2 mM) was used as indicator, working at the absorbance maximum of 557 nm with 10 mM Hepes (pH 7.5) as buffer and 0.1 M NaClO4 (for maintaining constant ionic strength), at 20 °C, following the CA-catalyzed CO2 hydration reaction for a period of 10–100 s (the uncatalyzed reaction needs around 60–100 s in the assay conditions, whereas the catalyzed ones are of around 6–10 s). The CO2 concentrations ranged from 1.7 to 17 mM for the determination of the kinetic parameters. The uncatalyzed rates were determined in the same manner and subtracted from the total observed rates. Enzyme concentrations in the assay system were about 10 nM for all the enzymes considered in the present study.
Effect of temperature on the NcoCA stability
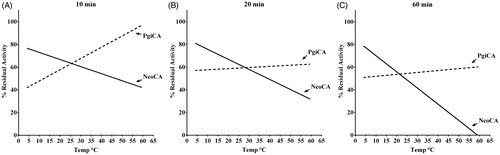
To compare the stability of NcoCA and PgiCA at different temperatures, enzyme solutions at the concentration of 3 μg/mL in 10 mM Tris/HCl, pH 8.3 were incubated at 4, 10, 25, 40, 50 and 60 °C for different times (10, 20, and 60 min). Enzyme aliquots (30 ng) were withdrawn at appropriate times and the residual activity was measured at 0 °C using CO2 as substrate.
PgiCA (γ-CA) preparation
PgiCA protein was prepared as described by Del Prete et al.Citation16.
Results
Sequence analysis
We aligned the amino acid sequence of NcoCA with the amino acid sequence of a mesophilic γ-CA, PgiCA, to try to identify unique features of this Antarctic enzyme (). The open reading frame of the CA gene identified in the genome of the Antarctic cyanobacterium encodes for polypeptide chain of 188 amino acid residues displaying 48% identity when compared with PgiCA (γ-CA from the bacterium Porphyromonas gingivalis). It may be observed that similar to the other investigated γ-CA, NcoCA has the conserved three His ligands, which coordinate the Zn(II) ion crucial for catalysis (His81, 117 and 122, CAM numbering systemCitation99,Citation100).
Figure 1. Amino acid sequence alignment of NcoCA and PgiCA (γ-CA). The metal ion ligands (His81, Hist117 and His122) are indicated in bold. The multialignment was performed with the program Muscle. Numbering system of the most known γ-CA, named CAM, was used Citation99,Citation100. The asterisk (*) indicates identity at all aligned positions; the symbol (:) relates to conserved substitutions, while (.) means that semi-conserved substitutions are observed.

Enzyme purification
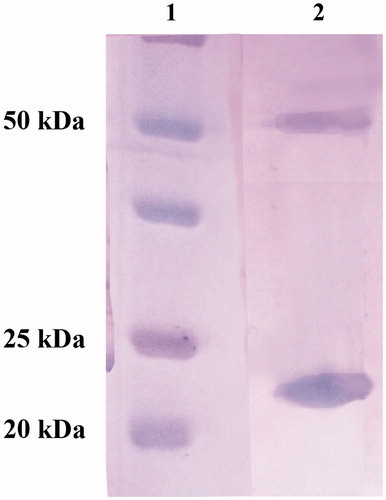
To study the biochemical properties of this psychrophilic enzyme, the recombinant NcoCA was isolated and purified to homogeneity at room temperature from E. coli (DE3) cell extract. Most of the CA activity was recovered in the soluble fraction of cell extract obtained after sonication and centrifugation. Using the affinity column (His-select HF Nickel affinity gel), NcoCA was purified to an apparent homogeneity of 90%, as indicated by SDS-PAGE (, lane 1). The apparent molecular weight estimated by SDS-PAGE of the purified His-tag NcoCA was 22.5 kDA. A subunit molecular mass of 22.0 kDa was calculated on the basis of the amino acid sequence translated from the NcoCA gene containing at the 5′ the His-tag nucleotide sequence. As described in the literature, tri-dimensional γ-CA structure revealed a trimeric arrangement for this class of CAs. Moreover, each monomer resembles an equilateral triangle stabilized by H bonds, salt bridges and hydrophobic interactionsCitation99,Citation100,Citation107. The SDS–PAGE of NcoCA showed two bands: one corresponding to a monomer with an apparent molecular weight of 22 kDa; while the second band showed an apparent molecular weight of 55 kDa, which corresponded to the trimeric form of the γ-CA, NcoCA. The trimer contains three active sites and each monomer contributes His residues located on the surface, at the interface between two adjacent monomers, to coordinate the Zn(II) ion crucial for catalysisCitation99,Citation100,Citation107–109.
Figure 2. SDS-PAGE of the recombinant NcoCA purified from E. coli cells. Legend: Lane 1, molecular markers; Lane 2, purified NcoCA from His-tag affinity column. NcoCA showed two bands: one corresponding to a monomer with an apparent molecular weight of 22 kDa; the second band showed an apparent molecular weight of 55 kDa.
Biochemical characterization
Protonography
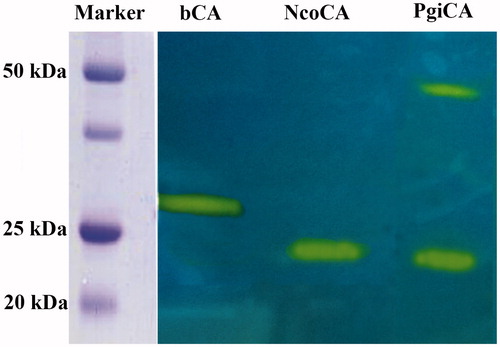
The hydratase activity of NcoCA on the SDS–PAGE gel was investigated and compared with that obtained for other CA classes, such as PgiCA (γ-CA) and bovine (b) bCA (α-CA). The gels were run under denaturing and non-reducing conditions. The protonogram showed in was obtained loading on the on the SDS–PAGE samples of NcoCA, PgiCA and bCA at 10 μg/well. As described in the experimental section, the protonography is based on monitoring the pH variation in the gel due to the CA-catalyzed conversion of CO2 to bicarbonate and protons. The protonogram is then stained with bromothymol blue, which is a widely used pH indicator. This dye appears blue in its deprotonated form, while its color changes to yellow in the protonated form. Thus, the production of ions (H+) during the CO2 hydration reaction, due to the CA hydratase activity, lowers the pH of the solution until the color transition point of the dye is reached, that is, at pH 6.8. It is interesting to note that NcoCA showed a different behavior on the protonogram respect to the other γ-CA, PgiCA (). The protonogram showed that PgiCA is present in two oligomeric state, monomer and trimer, while NcoCA showed only a hydratase activity band at the position corresponding to the molecular weight of its monomer. This suggests that probably the Antarctic enzyme reacted differently to the SDS present in the loading buffer with respect to the mesophilic enzyme, PgiCA. The α-CA, bCA, showed only one hydratase activity band (28 kDa) because its oligomeric state is monomericCitation22,Citation103,Citation104. It was possible to detect the hydratase activity of the inactive monomer of the γ-CA because, as described previously, following the electrophoresis, SDS was removed from the gel. This procedure potentially led to the rearrangement of γ-CA monomers in the gel, reconstituting thus the active trimeric form of the enzyme. The final result was the presence of a yellow band at the position of a γ-CA monomerCitation22,Citation103,Citation104.
Figure 3. Protonogram obtained using bCA (α-CA) and two γ-CAs: the Antarctic NcoCA and mesophilic PgiCA. The yellow bands correspond to the CA position on the gel responsible for the drop of pH from 8.2 to the transition point of the dye in the control buffer. Incubation time was of 20 s. NcoCA showed only one band (22 kDa), while PgiCA was present in two oligomeric states, the monomeric (22 kDa) and the trimeric (55 kDa) forms (see text for details).
Kinetic properties
A stopped-flow CO2 hydrase assay has been used to measure the catalytic activity of NcoCA. shows a comparison of the kinetic parameters for the CO2 hydration reaction catalyzed by NcoCA, hCAI and II (α-CA from Homo sapiens, isoform I and II, respectively), Can2 (β-CA from Cryptococcus neoformans), PgiCA (γ-CA from the bacterium Porphyromonas gingivalis) and CAM, (γ-CAs from Methanosarcina thermophila). It may be observed that NcoCA showed kinetic parameters about two times higher than the other γ-CA reported in (PgiCA). Moreover, the inhibition constant for acetazolamide (AZA) is similar to that obtained for the CAM.
Table 1. Kinetic parameters for the CO2 hydration reaction catalyzed by the α-, β- and γ-CAs at 20 °C and pH 7.5 in 10 mM HEPES buffer and 20 mM Na2SO4, measured at 20 °C, pH 8.3 in 20 mM TRIS buffer and 20 mM NaClO4Citation77.
Temperature effect on the stabilities of VchCA and bCA II
Stabilities of NcoCA and PgiCA were compared at the temperatures of 4, 10, 30, 40, 50, 60 °C (). After 10, 20 and 60 min of incubation time, the activities of NcoCA and PgiCA were determined using CO2 as substrate. As indicated in , after 60 min of incubation the activity of the Antarctic enzyme decreased to 20% at temperatures higher than 45 °C, and was completely inactivated at 60 °C. Otherwise, PgiCA, the mesophilic enzyme, retained its activity up to 60 °C () at all incubation times. It is interesting to note that the Antarctic enzyme showed an optimal activity at low temperature (4–20 °C) compared to the mesophilic counterpart (PgiCA). This behavior was observed at all the incubation times () reflecting a typical feature of the cold-adapted enzyme.
Discussion and conclusions
Antarctica can be considered as a huge natural laboratory for elucidating the evolutionary mechanisms developed by organisms, which were able to live and thrive in such an extreme habitat. Changes at the genetic level are relevant at the level of the molecular structure. For example, they might preserve the structure and function of an enzyme even though the molecule has to work in a completely different environment, i.e. the Antarctic sea where temperatures average −1.9 °C all year round. In general, cold-adapted enzymes show higher catalytic efficiency at low temperatures compared to their mesophilic counterparts. This characteristic is mainly due to the higher flexibility of the protein structure at low temperatures. Indeed, the tridimensional structures of the cold-adapted enzymes show a lower number of stabilizing factors, such as salt bonds, hydrogen bonds, hydrophobic interactions and a lower content of proline and arginine residues than their mesophilic counterpartsCitation77–80. For this reason, the study of CAs from Antarctic species is of interest, especially in relation to the strategies adopted by these organisms to achieve a normal level of catalytic activity at temperatures below that of the mesophilic species. The present paper describes the biochemical characterization of a γ-CA, named NcoCA, identified in the genome of the diazotrophic cyanobacteria belonging to the genus Nostoc, isolated from the freshwaters of Antarctic lakes, and confirms that all these adaptations to cold mentioned above are also present for this type of enzymes.
From the biochemical features of the purified enzyme and from the amino acid sequence analysis, NcoCA described in the present paper is a CA belonging to the γ-CA family. Enzyme from cold-adapted species commonly have an optimal activity at relatively low temperature; above this value, the loss of activity is usually attributed to thermal inactivation as a consequence of the increased structural flexibility of the protein. In fact, cold-adapted enzymes are typified by: (i) a higher catalytic efficiency than their mesophilic counterpart over a temperature range from 0 to 30 °C, and (ii) limited stability at moderate temperatures. It is immediately apparent that NcoCA meets the requirements of criterion (i) (at 25 °C NcoCAkcat/Km = 8.3 × 107, PgiCAkcat/Km = 5.4 × 107). Intriguingly, at 25 °C NcoCAkcat/Km is two order of magnitude higher than that determined for the γ-CA from the thermophilic bacteria Methanosarcina thermophila (CAMkcat/Km = 8.7 × 105). Even more clearly, it is evident that the Antarctic CA (NcoCA) completely meets the requirement of criteria (ii) (). NcoCA thermolability decreases with the increase of temperature (). The determinants of protein stability include structural factors, hydrophobic effects, and mainly weak interactions between atoms of the protein structureCitation110. Cold-adapted enzyme are characterized by clustering of glycine residues, providing local mobility; the disappearance of proline residues in loops, enhancing chain flexibility between secondary structures; a reduction in arginine residues which are capable of forming multiple salt bridges and H-bonds as well as a lower number of ion pairs, aromatic interactions, or H-bonds, compared to mesophilic enzymesCitation110. Comparison of the amino acid sequence of NcoCA with those of the mesophilic (PgiCA) or thermophilic (CAM) counterparts is far from being exhaustive, as it does not take into account all the details aforementioned. But the exploitation of recombinant DNA technology would allow the production of sufficient amount of NcoCA for structural studies to be undertaken by X-ray crystallography. In conclusion, the elucidation of the NcoCA structure, together with site-specific mutagenesis coupled with kinetic analysis, may provide a rational explanation for the thermolability displayed by the Antarctic CA with respect to the mesophilic and thermophilic such enzymes.
Declaration of interest
We thank the Distinguished Scientist Fellowship Program (DSFP) at KSU for funding this project. This work was also financed in part by an FP7 EU project (Dynano). The authors report no conflicts of interest.
References
- Migliardini F, De Luca V, Carginale V, et al. Biomimetic CO2 capture using a highly thermostable bacterial alpha-carbonic anhydrase immobilized on a polyurethane foam. J Enzyme Inhib Med Chem 2014;29:146–50
- Del Prete S, Vullo D, De Luca V, et al. Biochemical characterization of the delta-carbonic anhydrase from the marine diatom Thalassiosira weissflogii, TweCA. J Enzyme Inhib Med Chem 2014;29:906–11
- Del Prete S, De Luca V, Vullo D, et al. Biochemical characterization of the gamma-carbonic anhydrase from the oral pathogen Porphyromonas gingivalis, PgiCA. J Enzyme Inhib Med Chem 2014;29:532–7
- Capasso C, Supuran CT. Sulfa and trimethoprim-like drugs – antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J Enzyme Inhib Med Chem 2014;29:379–87
- Capasso C, Supuran CT. An overview of the selectivity and efficiency of the bacterial carbonic anhydrase inhibitors. Curr Med Chem 2015;22:2130–9
- Vullo D, Isik S, Del Prete S, et al. Anion inhibition studies of the alpha-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem Lett 2013;23:1636–8
- Monti SM, De Simone G, Dathan NA, et al. Kinetic and anion inhibition studies of a beta-carbonic anhydrase (FbiCA 1) from the C4 plant Flaveria bidentis. Bioorg Med Chem Lett 2013;23:1626–30
- Luca VD, Vullo D, Scozzafava A, et al. An alpha-carbonic anhydrase from the thermophilic bacterium Sulphurihydrogenibium azorense is the fastest enzyme known for the CO2 hydration reaction. Bioorg Med Chem 2013;21:1465–9
- Di Fiore A, Capasso C, De Luca V, et al. X-ray structure of the first `extremo-alpha-carbonic anhydrase', a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr D Biol Crystallogr 2013;69:1150–9
- Vullo D, De Luca V, Scozzafava A, et al. The first activation study of a bacterial carbonic anhydrase (CA). The thermostable alpha-CA from Sulfurihydrogenibium yellowstonense YO3AOP1 is highly activated by amino acids and amines. Bioorg Med Chem Lett 2012;22:6324–7
- Vullo D, De Luca V, Scozzafava A, et al. Anion inhibition studies of the fastest carbonic anhydrase (CA) known, the extremo-CA from the bacterium Sulfurihydrogenibium azorense. Bioorg Med Chem Lett 2012;22:7142–5
- Vullo D, Flemetakis E, Scozzafava A, et al. Anion inhibition studies of two alpha-carbonic anhydrases from Lotus japonicus, LjCAA1 and LjCAA2. J Inorg Biochem 2014;136:67–72
- Vullo D, Del Prete S, Osman SM, et al. Anion inhibition study of the beta-class carbonic anhydrase (PgiCAb) from the oral pathogen Porphyromonas gingivalis. Bioorg Med Chem Lett 2014;24:4402–6
- Del Prete S, Vullo D, Scozzafava A, et al. Cloning, characterization and anion inhibition study of the delta-class carbonic anhydrase (TweCA) from the marine diatom Thalassiosira weissflogii. Bioorg Med Chem 2014;22:531–7
- Del Prete S, Vullo D, Fisher GM, et al. Discovery of a new family of carbonic anhydrases in the malaria pathogen Plasmodium falciparum – the eta-carbonic anhydrases. Bioorg Med Chem Lett 2014;24:4389–96
- Del Prete S, Vullo D, De Luca V, et al. Biochemical characterization of recombinant beta-carbonic anhydrase (PgiCAb) identified in the genome of the oral pathogenic bacterium Porphyromonas gingivalis. J Enzyme Inhib Med Chem 2015;30:366–70
- Syrjanen L, Vermelho AB, Rodrigues Ide A, et al. Cloning, characterization, and inhibition studies of a beta-carbonic anhydrase from Leishmania donovani chagasi, the protozoan parasite responsible for leishmaniasis. J Med Chem 2013;56:7372–81
- Pan P, Vermelho AB, Capaci Rodrigues G, et al. Cloning, characterization, and sulfonamide and thiol inhibition studies of an alpha-carbonic anhydrase from Trypanosoma cruzi, the causative agent of Chagas disease. J Med Chem 2013;56:1761–71
- Del Prete S, Vullo D, De Luca V, et al. A highly catalytically active gamma-carbonic anhydrase from the pathogenic anaerobe Porphyromonas gingivalis and its inhibition profile with anions and small molecules. Bioorg Med Chem Lett 2013;23:4067–71
- Supuran CT, Capasso C. The eta-class carbonic anhydrases as drug targets for antimalarial agents. Expert Opin Ther Targets 2015;19:551–63
- De Simone G, Di Fiore A, Capasso C, Supuran CT. The zinc coordination pattern in the eta-carbonic anhydrase from Plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorg Med Chem Lett 2015;25:1385–9
- Del Prete S, De Luca V, Supuran CT, Capasso C. Protonography, a technique applicable for the analysis of eta-carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015. [Epub ahead of print]. doi:10.3109/14756366.2014.990963
- Capasso C, Supuran CT. An overview of the alpha-, beta- and gamma-carbonic anhydrases from Bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria? J Enzyme Inhib Med Chem 2015;30:325–32
- Del Prete S, De Luca V, Scozzafava A, et al. Biochemical properties of a new alpha-carbonic anhydrase from the human pathogenic bacterium, Vibrio cholerae. J Enzyme Inhib Med Chem 2014;29:23–7
- De Luca V, Vullo D, Scozzafava A, et al. Anion inhibition studies of an alpha-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Bioorg Med Chem Lett 2012;22:5630–4
- Zolfaghari ER, Syrjänen L, Barker H, et al. Drosophila melanogaster: a model organism for controlling Dipteran vectors and pests. J Enzyme Inhib Med Chem 2015;30:505–13
- Scozzafava A, Supuran CT. Glaucoma and the applications of carbonic anhydrase inhibitors. Subcell Biochem 2014;75:349–59
- McKenna R, Supuran CT. Carbonic anhydrase inhibitors drug design. Subcell Biochem 2014;75:291–323
- Carta F, Supuran CT, Scozzafava A. Sulfonamides and their isosters as carbonic anhydrase inhibitors. Future Med Chem 2014;6:1149–65
- Winum JY, Colinas PA, Supuran CT. Glycosidic carbonic anhydrase IX inhibitors: a sweet approach against cancer. Bioorg Med Chem 2013;21:1419–26
- Ward C, Langdon SP, Mullen P, et al. New strategies for targeting the hypoxic tumour microenvironment in breast cancer. Cancer Treat Rev 2013;39:171–9
- Supuran CT, Carta F, Scozzafava A. Metalloenzyme inhibitors for the treatment of Gram-negative bacterial infections: a patent review (2009–2012). Expert Opin Ther Pat 2013;23:777–88
- Supuran CT. Carbonic anhydrase inhibitors: an editorial. Expert Opin Ther Pat 2013;23:677–9
- Scozzafava A, Supuran CT, Carta F. Antiobesity carbonic anhydrase inhibitors: a literature and patent review. Expert Opin Ther Pat 2013;23:725–35
- Scozzafava A, Carta F, Supuran CT. Secondary and tertiary sulfonamides: a patent review (2008–2012). Expert Opin Ther Pat 2013;23:203–13
- Perez-Sayans M, Supuran CT, Pastorekova S, et al. The role of carbonic anhydrase IX in hypoxia control in OSCC. J Oral Pathol Med 2013;42:1–8
- Monti SM, Supuran CT, De Simone G. Anticancer carbonic anhydrase inhibitors: a patent review (2008–2013). Expert Opin Ther Pat 2013;23:737–49
- Masini E, Carta F, Scozzafava A, Supuran CT. Antiglaucoma carbonic anhydrase inhibitors: a patent review. Expert Opin Ther Pat 2013;23:705–16
- Harju AK, Bootorabi F, Kuuslahti M, et al. Carbonic anhydrase III: a neglected isozyme is stepping into the limelight. J Enzyme Inhib Med Chem 2013;28:231–9
- De Simone G, Alterio V, Supuran CT. Exploiting the hydrophobic and hydrophilic binding sites for designing carbonic anhydrase inhibitors. Expert Opin Drug Discov 2013;8:793–810
- Carta F, Supuran CT. Diuretics with carbonic anhydrase inhibitory action: a patent and literature review (2005–2013). Expert Opin Ther Pat 2013;23:681–91
- Capasso C, Supuran CT. Anti-infective carbonic anhydrase inhibitors: a patent and literature review. Expert Opin Ther Pat 2013;23:693–704
- Bertucci A, Moya A, Tambutte S, et al. Carbonic anhydrases in anthozoan corals – a review. Bioorg Med Chem 2013;21:1437–50
- Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov 2008;7:168–81
- Supuran CT. Carbonic anhydrases – an overview. Curr Pharm Des 2008;14:603–14
- Supuran CT. Diuretics: from classical carbonic anhydrase inhibitors to novel applications of the sulfonamides. Curr Pharm Des 2008;14:641–8
- Supuran CT. Development of small molecule carbonic anhydrase IX inhibitors. BJU Int 2008;101:39–40
- Zolnowska B, Slawinski J, Pogorzelska A, et al. Carbonic anhydrase inhibitors. Synthesis, and molecular structure of novel series N-substituted N'-(2-arylmethylthio-4-chloro-5-methylbenzenesulfonyl)guanidines and their inhibition of human cytosolic isozymes I and II and the transmembrane tumor-associated isozymes IX and XII. Eur J Med Chem 2014;71:135–47
- Slawinski J, Pogorzelska A, Zolnowska B, et al. Carbonic anhydrase inhibitors. Synthesis of a novel series of 5-substituted 2,4-dichlorobenzenesulfonamides and their inhibition of human cytosolic isozymes I and II and the transmembrane tumor-associated isozymes IX and XII. Eur J Med Chem 2014;82:47–55
- Slawinski J, Brzozowski Z, Zolnowska B, et al. Synthesis of a new series of N(4)-substituted 4-(2-aminoethyl)benzenesulfonamides and their inhibitory effect on human carbonic anhydrase cytosolic isozymes I and II and transmembrane tumor-associated isozymes IX and XII. Eur J Med Chem 2014;84:59–67
- Sethi KK, Verma SM, Tanc M, et al. Carbonic anhydrase inhibitors: synthesis and inhibition of the human carbonic anhydrase isoforms I, II, IX and XII with benzene sulfonamides incorporating 4- and 3-nitrophthalimide moieties. Bioorg Med Chem 2014;22:1586–95
- Scozzafava A, Passaponti M, Supuran CT, Gülçin I. Carbonic anhydrase inhibitors: guaiacol and catechol derivatives effectively inhibit certain human carbonic anhydrase isoenzymes (hCA I, II, IX and XII). J Enzyme Inhib Med Chem 2015;30:586–91
- Saluja AK, Tiwari M, Vullo D, Supuran CT. Substituted benzene sulfonamides incorporating 1,3,5-triazinyl moieties potently inhibit human carbonic anhydrases II, IX and XII. Bioorg Med Chem Lett 2014;24:1310–14
- Nasr G, Cristian A, Barboiu M, et al. Carbonic anhydrase inhibitors. Inhibition of human cytosolic isoforms I and II with (reduced) Schiff's bases incorporating sulfonamide, carboxylate and carboxymethyl moieties. Bioorg Med Chem 2014;22:2867–74
- Ghorab MM, Alsaid MS, Ceruso M, et al. Carbonic anhydrase inhibitors: synthesis, molecular docking, cytotoxic and inhibition of the human carbonic anhydrase isoforms I, II, IX, XII with novel benzenesulfonamides incorporating pyrrole, pyrrolopyrimidine and fused pyrrolopyrimidine moieties. Bioorg Med Chem 2014;22:3684–95
- Dekaminaviciute D, Kairys V, Zilnyte M, et al. Monoclonal antibodies raised against 167–180 aa sequence of human carbonic anhydrase XII inhibit its enzymatic activity. J Enzyme Inhib Med Chem 2014;29:804–10
- Davis RA, Vullo D, Supuran CT, Poulsen SA. Natural product polyamines that inhibit human carbonic anhydrases. Biomed Res Int 2014;2014:374079
- D'Ascenzio M, Carradori S, Secci D, et al. Selective inhibition of human carbonic anhydrases by novel amide derivatives of probenecid: synthesis, biological evaluation and molecular modelling studies. Bioorg Med Chem 2014;22:3982–8
- Ceruso M, Antel S, Vullo D, et al. Inhibition studies of new ureido-substituted sulfonamides incorporating a GABA moiety against human carbonic anhydrase isoforms I–XIV. Bioorg Med Chem 2014;22:6768–75
- Alterio V, Pan P, Parkkila S, et al. The structural comparison between membrane-associated human carbonic anhydrases provides insights into drug design of selective inhibitors. Biopolymers 2014;101:769–78
- Abdelrahim MY, Tanc M, Winum JY, et al. Dominant behaviors in the expression of human carbonic anhydrase hCA I activity. Chem Commun (Camb) 2014;50:8043–6
- Tarko L, Supuran CT. QSAR studies of sulfamate and sulfamide inhibitors targeting human carbonic anhydrase isozymes I, II, IX and XII. Bioorg Med Chem 2013;21:1404–9
- Slawinski J, Szafranski K, Vullo D, Supuran CT. Carbonic anhydrase inhibitors. Synthesis of heterocyclic 4-substituted pyridine-3-sulfonamide derivatives and their inhibition of the human cytosolic isozymes I and II and transmembrane tumor-associated isozymes IX and XII. Eur J Med Chem 2013;69:701–10
- Sethi KK, Vullo D, Verma SM, et al. Carbonic anhydrase inhibitors: synthesis and inhibition of the human carbonic anhydrase isoforms I, II, VII, IX and XII with benzene sulfonamides incorporating 4,5,6,7-tetrabromophthalimide moiety. Bioorg Med Chem 2013;21:5973–82
- Said HM, Hagemann C, Carta F, et al. Hypoxia induced CA9 inhibitory targeting by two different sulfonamide derivatives including acetazolamide in human glioblastoma. Bioorg Med Chem 2013;21:3949–57
- Pinard MA, Boone CD, Rife BD, et al. Structural study of interaction between brinzolamide and dorzolamide inhibition of human carbonic anhydrases. Bioorg Med Chem 2013;21:7210–15
- Nishimori I, Vullo D, Minakuchi T, et al. Restoring catalytic activity to the human carbonic anhydrase (CA) related proteins VIII, X and XI affords isoforms with high catalytic efficiency and susceptibility to anion inhibition. Bioorg Med Chem Lett 2013;23:256–60
- Leitans J, Sprudza A, Tanc M, et al. 5-Substituted-(1,2,3-triazol-4-yl)thiophene-2-sulfonamides strongly inhibit human carbonic anhydrases I, II, IX and XII: solution and X-ray crystallographic studies. Bioorg Med Chem 2013;21:5130–8
- Guzel-Akdemir O, Biswas S, Lastra K, et al. Structural study of the location of the phenyl tail of benzene sulfonamides and the effect on human carbonic anhydrase inhibition. Bioorg Med Chem 2013;21:6674–80
- Demirdag R, Yerlikaya E, Senturk M, et al. Heavy metal ion inhibition studies of human, sheep and fish alpha-carbonic anhydrases. J Enzyme Inhib Med Chem 2013;28:278–82
- Del Giudice R, Monti DM, Truppo E, et al. Human carbonic anhydrase VII protects cells from oxidative damage. Biol Chem 2013;394:1343–8
- Davis RA, Vullo D, Maresca A, et al. Natural product coumarins that inhibit human carbonic anhydrases. Bioorg Med Chem 2013;21:1539–43
- Ceruso M, Vullo D, Scozzafava A, Supuran CT. Inhibition of human carbonic anhydrase isoforms I-XIV with sulfonamides incorporating fluorine and 1,3,5-triazine moieties. Bioorg Med Chem 2013;21:6929–36
- Biswas S, McKenna R, Supuran CT. Effect of incorporating a thiophene tail in the scaffold of acetazolamide on the inhibition of human carbonic anhydrase isoforms I, II, IX and XII. Bioorg Med Chem Lett 2013;23:5646–9
- Alp C, Maresca A, Alp NA, et al. Secondary/tertiary benzenesulfonamides with inhibitory action against the cytosolic human carbonic anhydrase isoforms I and II. J Enzyme Inhib Med Chem 2013;28:294–8
- Akdemir A, Guzel-Akdemir O, Scozzafava A, et al. Inhibition of tumor-associated human carbonic anhydrase isozymes IX and XII by a new class of substituted-phenylacetamido aromatic sulfonamides. Bioorg Med Chem 2013;21:5228–32
- De Luca V, Maria G, De Mauro G, et al. Aspartic proteinases in Antarctic fish. Mar Genomics 2009;2:1–10
- Brier S, Maria G, Carginale V, et al. Purification and characterization of pepsins A1 and A2 from the Antarctic rock cod Trematomus bernacchii. FEBS J 2007;274:6152–66
- Capasso C, Carginale V, Scudiero R, et al. Phylogenetic divergence of fish and mammalian metallothionein: relationships with structural diversification and organismal temperature. J Mol Evol 2003;57 Suppl 1:S250–7
- Capasso C, Carginale V, Crescenzi O, et al. Solution structure of MT_nc, a novel metallothionein from the Antarctic fish Notothenia coriiceps. Structure 2003;11:435–43
- Capasso C, Abugo O, Tanfani F, et al. Stability and conformational dynamics of metallothioneins from the Antarctic fish Notothenia coriiceps and mouse. Proteins 2002;46:259–67
- D'Auria S, Carginale V, Scudiero R, et al. Structural characterization and thermal stability of Notothenia coriiceps metallothionein. Biochem J 2001;354:291–9
- Capasso C, Lees WE, Capasso A, et al. Cathepsin D from the liver of the Antarctic icefish Chionodraco hamatus exhibits unusual activity and stability at high temperatures. Biochim Biophys Acta 1999;1431:64–73
- Capasso C, Riggio M, Scudiero R, et al. Molecular cloning and sequence determination of a novel aspartic proteinase from Antarctic fish. Biochim Biophys Acta 1998;1387:457–61
- Carginale V, Trinchella F, Capasso C, et al. Gene amplification and cold adaptation of pepsin in Antarctic fish: a possible strategy for food digestion at low temperature. Gene 2004;336:195–205
- Carginale V, Capasso C, Scudiero R, Parisi E. Identification of cadmium-sensitive genes in the Antarctic fish Chionodraco hamatus by messenger RNA differential display. Gene 2002;299:117–24
- Scudiero R, Carginale V, Capasso C, et al. Structural and functional analysis of metal regulatory elements in the promoter region of genes encoding metallothionein isoforms in the Antarctic fish Chionodraco hamatus (icefish). Gene 2001;274:199–208
- Bargelloni L, Scudiero R, Parisi E, et al. Metallothioneins in antarctic fish: evidence for independent duplication and gene conversion. Mol Biol Evol 1999;16:885–97
- De Simone G, Monti SM, Alterio V, et al. Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense. Bioorg Med Chem Lett 2015;25:2002–6
- De Luca V, Del Prete S, Carginale V, et al. A failed tentative to design a super carbonic anhydrase having the biochemical properties of the most thermostable CA (SspCA) and the fastest (SazCA) enzymes. J Enzyme Inhib Med Chem 2015. [Epub ahead of print]. doi:10.3109/14756366.2014.1002403
- Alafeefy AM, Abdel-Aziz HA, Vullo D, et al. Inhibition of carbonic anhydrases from the extremophilic bacteria Sulfurihydrogenibium yellostonense (SspCA) and S. azorense (SazCA) with a new series of sulfonamides incorporating aroylhydrazone-, [1,2,4]triazolo[3,4-b][1,3,4]thiadiazinyl- or 2-(cyanophenylmethylene)-1,3,4-thiadiazol-3(2H)-yl moieties. Bioorg Med Chem 2014;22:141–7
- Vullo D, Luca VD, Scozzafava A, et al. The alpha-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1 is highly susceptible to inhibition by sulfonamides. Bioorg Med Chem 2013;21:1534–8
- Vullo D, De Luca V, Scozzafava A, et al. The extremo-alpha-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium azorense is highly inhibited by sulfonamides. Bioorg Med Chem 2013;21:4521–5
- Russo ME, Olivieri G, Capasso C, et al. Kinetic study of a novel thermo-stable alpha-carbonic anhydrase for biomimetic CO2 capture. Enzyme Microb Technol 2013;53:271–7
- Capasso C, De Luca V, Carginale V, et al. Biochemical properties of a novel and highly thermostable bacterial alpha-carbonic anhydrase from Sulfurihydrogenibium yellowstonense YO3AOP1. J Enzyme Inhib Med Chem 2012;27:892–7
- Vullo D, De Luca V, Del Prete S, et al. Sulfonamide inhibition studies of the gamma-carbonic anhydrase from the Antarctic cyanobacterium Nostoc commune. Bioorg Med Chem 2015;23:1728–34
- De Luca V, Del Prete S, Carginale V, et al. Cloning, characterization and anion inhibition studies of a gamma-carbonic anhydrase from the Antarctic cyanobacterium Nostoc commune. Bioorg Med Chem Lett 2015. [Epub ahead of print]. doi: 10.1016/j.bmcl.2015.03.010
- Johnson M, Zaretskaya I, Raytselis Y, et al. NCBI BLAST: a better web interface. Nucleic Acids Res 2008;36:W5–9
- Zimmerman SA, Tomb JF, Ferry JG. Characterization of CamH from Methanosarcina thermophila, founding member of a subclass of the {gamma} class of carbonic anhydrases. J Bacteriol 2010;192:1353–60
- Ferry JG. The gamma class of carbonic anhydrases. Biochim Biophys Acta 2010;1804:374–81
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32:1792–7
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680–5
- Del Prete S, De Luca V, Iandolo E, et al. Protonography, a powerful tool for analyzing the activity and the oligomeric state of the gamma-carbonic anhydrase identified in the genome of Porphyromonas gingivalis. Bioorg Med Chem 2015;23:3747–50
- De Luca V, Del Prete S, Supuran CT, Capasso C. Protonography, a new technique for the analysis of carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015;30:277–82
- Chirica LC, Elleby B, Jonsson BH, Lindskog S. The complete sequence, expression in Escherichia coli, purification and some properties of carbonic anhydrase from Neisseria gonorrhoeae. Eur J Biochem 1997;244:755–60
- Khalifah RG. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J Biol Chem 1971;246:2561–73
- Zimmerman SA, Ferry JG, Supuran CT. Inhibition of the archaeal beta-class (Cab) and gamma-class (Cam) carbonic anhydrases. Curr Top Med Chem 2007;7:901–8
- Innocenti A, Zimmerman SA, Scozzafava A, et al. Carbonic anhydrase activators: activation of the archaeal beta-class (Cab) and gamma-class (Cam) carbonic anhydrases with amino acids and amines. Bioorg Med Chem Lett 2008;18:6194–8
- Zimmerman S, Innocenti A, Casini A, et al. Carbonic anhydrase inhibitors. Inhibition of the prokariotic beta and gamma-class enzymes from Archaea with sulfonamides. Bioorg Med Chem Lett 2004;14:6001–6
- Feller G. Psychrophilic enzymes: from folding to function and biotechnology. Scientifica (Cairo) 2013;2013:512840