Abstract
The type II transmembrane serine protease matriptase is a potential target for anticancer therapy and might be involved in cartilage degradation in osteoarthritis or inflammatory skin disorders. Starting from previously described nonspecific thrombin and factor Xa inhibitors we have prepared new noncovalent substrate-analogs with superior potency against matriptase. The most suitable compound 35 (H-d-hTyr-Ala-4-amidinobenzylamide) binds to matriptase with an inhibition constant of 26 nM and has more than 10-fold reduced activity against thrombin and factor Xa. The crystal structure of inhibitor 35 was determined in the surrogate protease trypsin, the obtained complex was used to model the binding mode of inhibitor 35 in the active site of matriptase. The methylene insertion in d-hTyr and d-hPhe increases the flexibility of the P3 side chain compared to their d-Phe analogs, which enables an improved binding of these inhibitors in the well-defined S3/4 pocket of matriptase. Inhibitor 35 can be used for further biochemical studies with matriptase.
Introduction
Matriptase is one of the best characterized members among the 17 conserved type II transmembrane serine proteases (TTSPs) found in human and miceCitation1. Like all other TTSPs matriptase possesses a complex modular structure containing an extracellular C-terminal trypsin-like serine protease domain. It is widely expressed in epithelial tissues and mediates various pleiotropic effects in development, cell–cell adhesion and tissue homeostasisCitation2. Matriptase is required for normal epithelial development and postnatal survival, hence, knockout mice die within 48 h of birth due to severe dehydration caused by impaired epidermal barrier functionCitation3. Individuals harboring a missense mutation leading to a Gly→Arg change in residue 216 of the protease domain (chymotrypsin numbering), which results in strongly reduced matriptase activity, suffer from ichthyosis with hypotricosisCitation4,Citation5. Furthermore, matriptase mRNA and protein are downregulated in inflamed colonic tissues from Crohn’s disease and ulcerative colitis patients. The loss of matriptase leads to leaky tight junctions within the gut and reduced transepithelial resistance promoting intestinal inflammationCitation6,Citation7.
Beside these important functions in normal physiological processes matriptase can activate numerous substrates involved in cancerCitation8 and other diseases. For instance, matriptase efficiently cleaves soluble single-chain pro-urokinase type plasminogen activator (pro-uPA), as well as receptor-bound pro-uPACitation9–11. uPA promotes extracellular proteolysis by generating the broad spectrum protease plasmin, which is an activator of MMPs, of certain growth hormones, and is involved in the degradation of extracellular matrix proteins (ECM). The components of the plasminogen activation system are upregulated in several tumor entities and promote tumor invasion and metastasis. In addition to some other proteases including hepatocyte growth factor activator (HGFA), matriptase efficiently converts pro-hepatocyte growth factor/scattering factor (pro-HGF/SF) to its active formCitation10. HGF/SF has high affinity to the receptor tyrosine kinase c-Met, thereby inducing signaling pathways, which promote tumorigenesis and invasive growthCitation12,Citation13. The G-protein coupled protease-activated receptor 2 (PAR2) is an additional candidate substrate of matriptase, which is localized on the extracellular surfaceCitation9. PAR2 mediates cell adhesion, cell motility and inflammation and has been suggested to stimulate metastasisCitation14. Beside these functions in cancer, a matriptase-catalyzed PAR2 activation also leads to an increased collagenase expression contributing to enhanced cartilage degradation in osteoarthritisCitation15. Moreover, a strong increase in matriptase expression was observed in inflammatory skin disordersCitation16. Recent findings suggest that the host protease matriptase might also be involved in the activation of certain H1 and H9 influenza virus hemagglutinins, which is essential for virus propagationCitation17,Citation18.
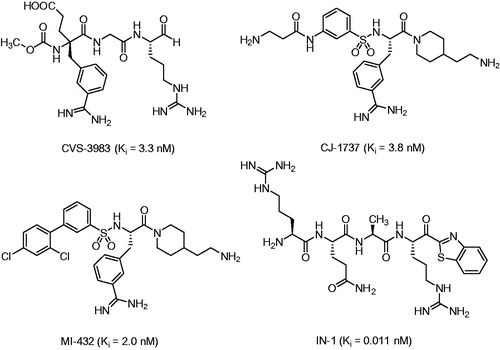
Therefore, matriptase emerged as a potential drug target, especially for the treatment of epithelial tumors. First successful proof of concepts studies with prostate cancer models in mice have been performed with the arginal-derived matriptase inhibitor CVS-3983Citation19, and with the 3-amidinophenylalanine derivative CJ-1737Citation20 (). Moreover, CJ-1737 and related inhibitors reduced the pro-HGF/SF driven phosphorylation of the HGF receptor/c-Met and the overall cellular invasiveness of the human pancreatic adenocarcinoma cell line AsPC-1Citation21. Even stronger effects on pro-HGF/SF induced c-Met activation on primary mammary carcinoma cells and three other human breast cancer cell lines were described for the ketobenzothiazole inhibitor IN-1Citation22 (), meanwhile various highly potent ketone-derived inhibitors have been describedCitation23,Citation24. The invasiveness and migration of various tumor cell lines could be also inhibited by other benzamidine-derived matriptase inhibitorsCitation25,Citation26.
During the search for inhibitors of matriptase-2, a related TTSP, we have previously observed a significant inhibitory potency of the substrate analog inhibitor BAPA (benzylsulfonyl-d-Arg-Pro-4-amidinobenzylamide, compound 5 in ) against matriptase (Ki = 55 nM)Citation27. Its C-terminal arginine mimetic 4-amidinobenzylamide (4-Amba) is an excellent anchor for all trypsin-like serine proteases and can be produced in large quantitiesCitation28, but it does not contribute to any selectivity within this family. It was initially described during the development of the thrombin inhibitor melagatran/ximelagatranCitation29 and was later used for the design of potent uPA, factor Xa, factor VIIa, and plasmin inhibitorsCitation30–33.
Because of the poor selectivity profile of BAPA, which is a significant stronger inhibitor of the clotting proteases thrombin and factor Xa (fXa) with inhibition constants of 3.5 and 2.5 nM, respectivelyCitation34, we tried to develop new substrate-analog inhibitors with improved selectivity for matriptase. The results of this work are summarized in the current paper.
Materials and methods
Reagents for synthesis, including protected standard amino acids, coupling reagents and solvents were obtained from Bachem, Fluka, Acros, Merck or Aldrich.
Analytical HPLC experiments were performed on a Shimadzu LC-10A system (column: Nucleodur C18, 5 μm, 100 Å, 4.6 × 250 mm, Machery-Nagel, Düren, Germany) with a linear gradient of acetonitrile (solvent B) and water (solvent A) both containing 0.1% TFA at a flow rate of 1 ml/min (1% increase solvent B per min, starting at 1% solvent B), detection at 220 nm. The final inhibitors were purified to more than 95% using preparative HPLC (pumps: Varian PrepStar Model 218 gradient system, detector: ProStar Model 320 with detection at 220 nm, fraction collector: Varian Model 701; column: C8, Nucleodur, 5 μm, 100 Å, 32 × 250 mm, Macherey-Nagel, Düren, Germany) by a linear gradient with the same solvents as described above at a flow rate of 20 ml/min. All final inhibitors were obtained as TFA-salts after lyophilization. The molecular mass of the synthesized compounds was determined using a QTrap 2000 ESI spectrometer (Applied Biosystems, now Life Technologies, Carlsbad, CA). The 1H- and 13C-NMR spectra were recorded on a Jeol JNM-GX-400 at 400 and 100 MHz (Jeol Inc., Peabody, MA) and are referenced to internal solvent signals.
Synthesis
The most selective inhibitor 35 was synthesized as described in Scheme 1.
Scheme 1. Synthesis of inhibitor 35. Conditions and reagents (a) 1 equiv Boc-Ala-OH, 1 equiv BOP, 3 equiv DIPEA; (b) TFA, 1 h room temperature; (c) 1.15 equiv Boc2O in dioxane and water, pH ∼ 8.5 adjusted with 1 N NaOH; (d) 1 equiv BOP, 3 equiv DIPEA; (e) TFA, 1 h, room temperature, preparative HPLC.
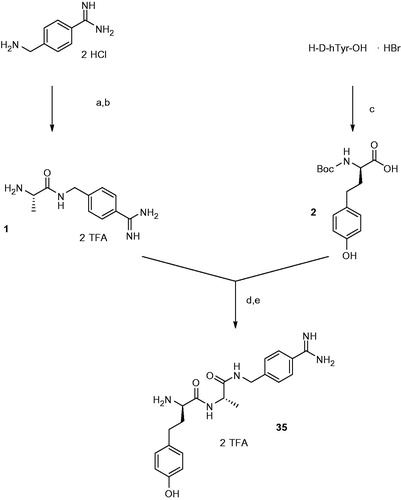
H-Ala-4-Amba · 2 TFA (1)
Boc-Ala-OH (333 mg, 1.76 mmol) and 4-amidinobenzylamine · 2 HClCitation35 (391 mg, 1.76 mmol) were suspended in 10 ml DMF. After cooling to 0° C the mixture was treated with 1.155 g (2.61 mmol) BOP and 1.36 ml (7.83 mmol) DIPEA. After stirring overnight at room temperature, the solvent was removed in vacuo, and the Boc-protected intermediate was purified by preparative HPLC. The product containing fractions (HPLC: tR = 23.89 min, start at 1% solvent B) were combined and evaporated. The remaining residue was treated with 10 ml trifluoroacetic acid and stirred for one hour at room temperature. The solvent was evaporated and the product was lyophilized from water providing a white solid (yield: 285 mg, 0.64 mmol, 36%, HPLC: tR = 8.88 min, start at 1% solvent B, MS calcd m/z 220.13, m/z found 221.2 (M + H)+. 1H NHMR (400 MHz, DMSO-d6) 1.39 (d, J = 7.20 Hz, 3H), 3.91 (s, 1H), 4.45 (d, J = 6.00 Hz, 2H), 7.49 (d, J = 8.00 Hz, 2H), 7.79 (d, J = 8.00 Hz, 2H), 8.10 (s, 3H), 8,98 (t, J = 6.00 Hz, 1H), 9,26 (s, 1H).
Boc-d-hTyr-OH (2)
111 mg (0.4 mmol) of H-d-hTyr-OH· HBr (Iris Biotech GmbH, Marktredwitz, Germany) was dissolved in 4 ml dioxane, 2 ml water and 1 ml 1 N NaOH. The mixture was cooled to 0° C and treated with 100 mg (0.46 mmol) Boc2O. After 10 min the ice bath was removed and the mixture was stirred at room temperature for 2 h. The solvent was removed in vacuo and the remaining residue was dissolved in a mixture of 5% KHSO4 and ethyl acetate. The organic layer was washed 3 × with 5% KHSO4 and 3 × with saturated aqueous NaCl. The organic phase was dried over Na2SO4, and the solvent was removed in vacuo to afford a colorless viscous oil (yield: 95 mg, 0.322 mmol, 80.5%, HPLC start at 1% solvent B: tR = 40.32 min, MS calcd m/z 295.14, m/z found 294.25 (M−H)−. 1H NHMR (400 MHz, DMSO-d6) 1.39 (s, 9H), 1.83 (m, 2 h), 2.50 (m, 2 h), 3.82 (m, 1H), 6.65 (d, J = 8.40 Hz, 2H), 6.96 (d, J = 8.40 Hz, 2H), 7.21 (m, 1H) 9.11 (s, 1 H), 12.40 (s, 1H).
H-d-hTyr-Ala-4-Amba · 2 TFA (35)
H-Ala-4-Amba · 2 TFA (111 mg, 0.25 mmol) and Boc-d-hTyr-OH (74 mg, 0.25 mmol) were suspended in 5 mL of DMF. The mixture was cooled to 0° C and treated with 110 mg (0.25 mmol) BOP and 130 μL (0.75 mmol) DIPEA. After stirring overnight at room temperature, the solvent was removed in vacuo. The remaining residue was treated with 5 ml trifluoroacetic acid and stirred for one hour at room temperature. After precipitation in ether and centrifugation, the obtained solid was purified by preparative HPLC. The product containing fractions were combined, concentrated and lyophilized from water providing a white solid (yield: 32 mg, 0.05 mmol, 20%, HPLC: tR = 17.33 min, start at 1% solvent B, MS calcd m/z 397.48, m/z found 398.21(M + H)+ 1H NHMR (400 MHz, DMSO-d6) 1.29 (d, J = 6.16 Hz, 3H), 1.94 (m, 2H), 3.92 (m, 1H), 4.40 (m, 3H), 6.69 (d, J = 8.80 Hz, 2H), 6.96 (d, J = 8.40 Hz, 2H), 7.46 (d, J = 8.80 Hz, 2H), 7.77 (d, J = 8.40 Hz, 2H), 8.18 (s, 3H), 8.69 (m, 1H), 8,76 (m, 1H), 9.21 (s, 3), 9.26 (s 1H 13C NMR (100 MHz, DMSO-d6) 19.0, 30.1, 34.0, 42.3, 49.1, 52.8, 115.8, 127.1, 127.8, 128.7, 129.5, 131.1, 146.3, 156.2, 166.1, 168.7, 172.4).
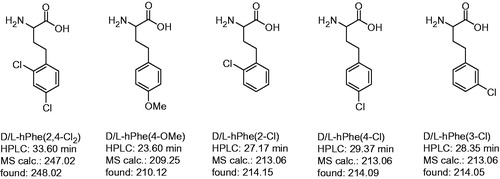
Racemic P3 homoamino acids, which were not commercially available (), were prepared from appropriately substituted phenylethyl chlorides or bromides by reaction with ethyl acetamidocyanoacetate in presence of potassium carbonate and potassium iodide, as described previouslyCitation36. The synthesis of the inhibitors with these residues was performed as shown in Scheme 1 for the d-hTyr inhibitor. The final diastereomeric inhibitors could be separated by preparative reversed phase HPLC. Substrate analog inhibitors of trypsin-like serine proteases normally prefer d-configurated P3-residues. Therefore, the compounds with stronger inhibitory potency were assigned to contain the P3 residue in d-configuration, whereas their analogs with lower potency possess a l-configurated P3 amino acid.
Figure 2. Analytical data and abbreviations of prepared racemic homo amino-acids. Their determined mass corresponds to the (M + H)+ ion, the HPLC analysis started at 1% solvent B.
For the synthesis of the benzylsulfonyl protected inhibitors shown in , the appropriate benzylsulfonyl-protected P3 amino acids were used instead of the Boc derivatives. The analytical data of all inhibitors are provided as Supplementary material available online.
Determination of inhibition constants
The Ki values for the inhibition of the catalytic domain of matriptase (23 pM in assay), which was prepared as described previouslyCitation20, and factor Xa (97 pM in assay, purchased from Enzyme Research South Bend, Indiana, USA) were determined with the substrate methylsulfonyl-d-Arg-Pro-Arg-AMC (prepared in houseCitation37, for matriptase: KM = 6 μM, used concentrations in assay 14, 7 and 3.5 μM; for FXa: KM = 28 μM, used concentrations in assay 50, 25 and 12.5 μM), while Tos-Gly-Pro-Arg-AMC (KM = 5.4 μM, used concentrations in assay 10, 5 and 2.5 μM) was used for bovine thrombin prepared according to WalsmannCitation38 (31 pM in assay). The measurements were performed in a Fluoroscan microplate reader (λex = 355 and λem = 460 nm; Thermo Fisher Scientific, Vantaa, Finland) with 100 μl buffer (50 mM Tris, 154 mM NaCl, pH 8.0), 20 μl substrate solution (substrate dissolved in water) and 20 μl enzymeCitation39. The Ki values were obtained from Dixon plotsCitation40 and are the average of at least two measurements.
Crystal structure analysis
Cocrystallization of trypsin with inhibitor 35: Bovine beta-Trypsin (Sigma, # T8003) was dissolved in a solution of 10 mM CaCl2 at 20 mg/ml for 30 minutes on ice. The inhibitor 35 was dissolved in water at 2.5 mg/ml. The crystallisation buffer contained 0.1 M imidazole, 0.1 M (NH4)2SO4, 20% PEG 8000, 0.1% sodium azide at pH 8. In the next step, 50 μL of the trypsin solution was mixed with 10 μL of the inhibitor solution and filled up to 100 μL with water. This new solution was mixed 1:1 with the crystallization buffer and 5 μL of this solution were placed in the center of a cover slip. Crystallization was carried out at 16 °C by the hanging-drop method. The wells of the crystallization trays were filled with 500 μL of the crystallization buffer. Subsequently, the cover slips were placed over the wells and sealed. Crystals of good diffracting quality could be obtained after 20 to 30 days.
Data collection and processing: Obtained crystals were frozen at 77 K for data collection. The data set was collected at 100 K at synchrotron beamline 14.2 at the BESSY using MARMOSAIC 225 mm CCD detector. Data processing and scaling were performed using the HKL2000 packageCitation41.
Structure determination and refinement: The coordinates of bovine trypsin (PDB: 2ZFS)Citation42, were used for molecular replacement with Phaser from the CCP4 program packageCitation43. For initial rigid body refinement of the protein molecule, followed by repeated cycles of maximum likelihood energy minimization simulated annealing and B-factor refinement the program PHENIXCitation44 was used. A randomly chosen 5% of all data were used for the calculation of Rfree and were not used in the refinement. Amino acid side chains were fit into σ-weighted 2Fo – Fc and Fo – Fc electron density maps using CootCitation45. After the first refinement cycle, water molecules and subsequently ions and ligands were located in the electron density and added to the model. Restraints were applied to bond lengths and angles, planarity of aromatic rings and van der Waals contacts. Multiple side-chain conformations were built in case an appropriate electron density was observed and maintained during the refinement, and if the minor populated side chain showed at least 20% occupancy. The final model was validated using PHENIX own validation options or ADIT. Data collection, unit cell parameters and refinement statistics are given in . The naming of the protein amino acids was done according to Bode et al.Citation46. Coordinates and structure factors have been deposited in the Protein Data Bank with the accession code 4MTB.
Table 1. Data collection and refinement statistics for the trypsin/inhibitor 35 complex.
Table 2. Inhibition of matriptase, thrombin and fXa by benzylsulfonyl-protected inhibitors available from previous workCitation27,Citation49,Citation50.
Results
Modeled complex of BAPA in matriptase
Due to the limited amount of matriptase we could not determine its crystal structure in complex with substrate analog inhibitors so far. For initial modeling of their binding mode in matriptase we used the recently described crystal structure of a trypsin mutant in complex with benzylsulfonyl-d-Arg-Gly-4-Amba, the P2 glycine analog of BAPA 5 (3pmj.pdb)Citation47. The complex was superimposed by a Cα-atom fit with the previously determined crystal structure of matriptase (2gv6.pdb)Citation20, all water molecules were deleted. After replacement of the P2 glycine with proline, the bound inhibitor was energy-minimized using the program MOECitation48, whereas the coordinates of matriptase were kept constant.
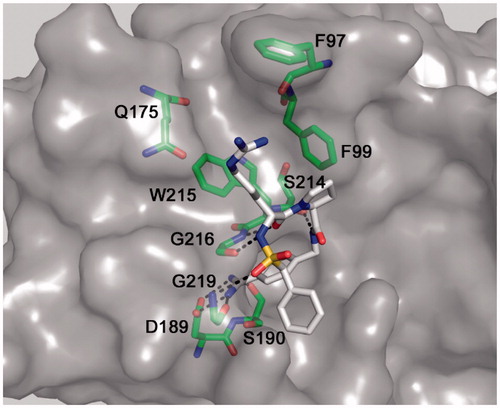
The obtained model reveals the expected overall binding mode known from many structures of various trypsin-like serine proteases in complex with substrate analog inhibitors, containing a P3 residue in d-configuration (). The d-Arg side-chain is oriented towards the distal S3/4 binding pocket above Trp215 and might be involved in cation-π interactions. However, its side-chain guanidino group is not involved in any specific hydrogen bonds with matriptase. In contrast, all known interactions of the benzamidine group in the S1 pocket and between the inhibitor backbone and the matriptase residues Ser214, Gly216 and Gly219 were found. The model suggested that it should be possible to replace the P3 d-arginine and P2 proline by other residues to improve affinity of the inhibitors.
Figure 3. Modeled binding mode of BAPA (inhibitor 5) in matriptase. The coordinates of matriptase were taken from the pdb entry 2gv6. The protease is shown with its solvent-accessible transparent surface in gray. The inhibitor is displayed as stick model with carbon atoms in white, nitrogen in blue, oxygen in red, and sulfur in yellow. Selected residues of matriptase forming the S3/4 pocket (Trp215 at the bottom, Phe99 on the right, Gln175 on the left and Phe97 on the top), as well as residues involved in polar contacts to the inhibitor (Asp189, Ser190, Gly216, and Gly219) are labeled and shown as sticks with carbons in green. All colored figures were prepared using PyMOL v0.98 (DeLano Scientific, San Carlos, CA).
Determination of inhibition constants
Initially, benzylsulfonyl protected inhibitors containing d-configurated P3 residues available from previous studiesCitation27,Citation49,Citation50 were screened with matriptase, thrombin and fXa. In a first series, the P3 position was modified, whereas proline was maintained as P2 residue, while in a second one the d-arginine in P3 position was kept constant and the P2 residue was modified ().
Although a few analogs, e.g. inhibitors 3–6, revealed a significant matriptase affinity with inhibition constants < 50 nM, all compounds with proline in P2 position showed a considerably stronger thrombin inhibition and most substances also possess a high fXa affinity. Interestingly, a ∼25-fold improved matriptase inhibition was found for the d-homophenylalanine (d-hPhe) derivative 3 over its d-Phe analog 11. A relatively high potency was also determined for inhibitors containing basic P3 side chains, which are probably involved in cation-π-interactions to matriptase residues Trp215 and Phe99.
Therefore, we screened a second series 15–22 with d-Arg as constant P3 residue, which was available from previous work on inhibitors of the human airway trypsin-like protease (HAT)Citation49,Citation51. In comparison to the P2 Pro inhibitor 5, a 1.5-fold stronger matriptase inhibition was observed for the Ala-analog 15. As expected, this replacement strongly diminished the inhibition of thrombin, whereas the affinity against fXa was slightly enhanced. A similar matriptase inhibition was found for the P2 Arg inhibitor 16, all other P2 modifications reduced the potency.
From previous work we know that the potency of this inhibitor type against fXa strongly depends on the presence of a suitable P4 group. Elimination of the N-terminal sulfonyl group of Bzls-dSer(tBu)-Gly-4-Amba (Ki = 14 nM) resulted in a ≈1000-fold reduced fXa affinity (Ki = 16 μM), whereas the affinity against thrombin was only 6-fold reducedCitation52. Moreover, in previous work we found an enhanced matriptase inhibition with biphenyl-3-sulfonyl-substituted derivatives of 3-amidinophenylalanine. The most potent derivatives of this series were substituted by chlorine atoms (e.g., see inhibitor MI-432 in ) or methoxy groups at their N-terminal phenyl ring, which occupies a similar position in the S3/4 pocket of matriptase as known for the d-configurated P3 side chain of substrate-analog inhibitors. Based on these findings and due to the high potency of compound 3, new substrate analog inhibitors without a P4 benzylsulfonyl group in combination with chloro- or methoxy-substituted d-hPhe analogs in P3 position have been prepared. All these inhibitors were synthesized with Pro or Ala as P2 residue (). Comparison of compound 3 with compound 23 revealed that the deletion of the Bzls-group reduced the matriptase inhibition only by a factor of five, whereas the potency against fXa was ≈200-fold decreased. Further replacement of P2 Pro by Ala provided compound 36, the first substrate-analog inhibitor with superior affinity for matriptase compared to thrombin and fXa. However, all chloro- or methoxy-substituted d-hPhe analogs showed reduced matriptase affinity, whereas in few cases the inhibition of thrombin and fXa was slightly enhanced. Interestingly, incorporation of the commercially available d-hTyr provided inhibitor 35, which possesses an improved affinity against all three proteases and maintains the selectivity profile as found for compound 36. As expected, a poor inhibitory potency was found for all inhibitors containing l-configurated P3 amino acids.
Table 3. Inhibition of matriptase, thrombin and factor Xa by inhibitors lacking a P4 residue.
Crystal structure of inhibitor 35 in trypsin and model in complex with matriptase
Although we could reduce the potency against thrombin and fXa it was found that the d-hTyr inhibitor 35 and its d-hPhe analog 36 are very potent inhibitors of bovine trypsin with Ki values of 0.81 nM and 3.7 nM, respectively. Since we had insufficient amounts of matriptase for structure determination, inhibitor 35 was crystallized in the surrogate protease trypsin (). All typical contacts of the P1 benzamidine within the S1 pocket and from the backbone of the inhibitor to trypsin residues Ser214 and Gly216 have been found and are identical as previously observed in crystal structures of substrate-analog inhibitors with thrombinCitation50, fXaCitation52 or uPACitation30. The P2 carbonyl binds to the side-chain amide of Gln192 and two water molecules, one of them mediates a contact to the free P3 amino group. The d-hTyr backbone interacts with Gly216, and the P3 side chain fits well into the distal S3/4 pocket. The hydroxyl group forms a close contact to the carbonyl of the Gln175 side-chain amide and is surrounded by a complex water network. Only two of them, which directly bind to the inhibitor, are shown in , one of them enables water-mediated contacts to the main chain carbonyls of Thr98 and Gln175.
Figure 4. Compound 35 in complex with trypsin and matriptase: (A) Crystal structure of compound 35, shown as sticks (carbon in green, nitrogen in blue, oxygen in red, and water molecules as red spheres) in complex with bovine trypsin presented with its backbone in beige (4MTB.pdb). Selected trypsin residues are shown as sticks with light pink carbon atoms; (B) Model of compound 35 in the active site of matriptase (shown with gray surface and backbone as cartoon in green), obtained by superimposition of the 35/trypsin complex with the crystal structure of matriptase (2GV6), followed by an energy minimization of the d-homotyrosine side-chain using the program MOECitation48. All other inhibitor atoms and the matriptase residues were fixed, water molecules were deleted. A pdb file of the modeled matriptase/inhibitor 35 complex is provided as Supplementary material for download.
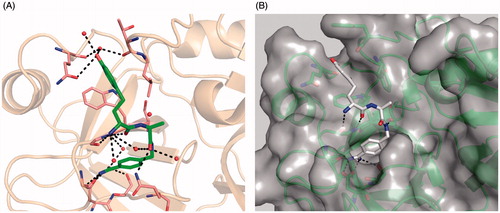
Superposition of the complex with the crystal structure of matriptase (2gv6.pdb) suggests a very similar binding mode, all characteristic interactions were maintained (). The positively charged amidine group binds to the side chains of matriptase residues Asp189, Ser190 and to the carbonyl oxygen of Gly219. Additional H-bonds exist between the P1 amide NH and the carbonyl oxygen of Ser214 and between the backbone of d-hTyr with the carbonyl oxygen and NH of Gly216. Moreover, the insertion of an additional methylene group increases the flexibility of d-hTyr, which enables a deeper binding of the P3 side chain into the distal S3/4 pocket of matriptase. This might explain the increased affinity of the d-hPhe and d-hTyr inhibitors compared to the less flexible d-Phe or d-Cha-analogs.
Discussion
Starting with a screening of available benzylsulfonyl-protected inhibitors we identified d-hPhe and Ala as most suitable P3 and P2 residues for substrate-analog matriptase inhibitors. The replacement of P2 Pro by Ala is a simple method to achieve a significant reduction in thrombin affinity, whereas it leads to a slightly better matriptase and fXa inhibition. A P2 Ala is also found in the P4-P4’ segment (RQAR↓VVGG) of the autocatalytic cleavage site of matriptase and it can be well accommodated in the S2 pocket below the side chains of Phe99 and His57. The RQAR sequence served recently for the design of highly potent arginyl-ketone derived matriptase inhibitorsCitation23. For comparison we have also incorporated the natural P4-P2 substrate segment in our series, the resulting inhibitor H-Arg-Gln-Ala-4-Amba possesses a ca. 5-fold reduced potency against matriptase (Ki = 0.12 μM) compared with inhibitor 35, although it has a nearly 100-fold and therefore more pronounced selectivity over thrombin (Ki = 14.7 μM) and fXa (Ki = 14.5 μM). Otherwise, l-configurated peptides can suffer from rapid degradation in vivo, whereas the incorporation of d-amino acids often contributes to an improved stability.
The affinity against fXa could be significantly reduced by deletion of the P4 benzylsulfonyl group. In case of fXa it binds in a shallow subpocket above the Cys220-Cys191 disulfide-bridge, where its methylene group comes in close contact with one of the phenyl-ring carbons of the P1 4-Amba residue (distance 3.7 Å), thus stabilizing the compact horseshoe-like inhibitor conformationCitation52. Although the binding mode of the benzylsulfonyl group in thrombin is very similarCitation53 and probably also comparable in matriptase (see model in ), its elimination has a less pronounced influence on the inhibition of these two enzymes.
The improved potency of the d-hPhe and d-hTyr inhibitors against matriptase resembles a trend, which has been previously observed during the development of substrate-analog fXa inhibitorsCitation31. In that case also the shorter P3 d-Phe provides less active inhibitors. The improved potency can be explained based on the crystal structure of inhibitor 35 in complex with trypsin, assuming a similar binding mode in fXa and matriptase. Due to the methylene insertion, the side chains of the P3 homoamino acids find better access to the well-defined S3/4 binding sites of fXa and matriptase. In trypsin, the d-hTyr enables additional water mediated contacts to carbonyl groups at the end of this pocket, although this results in only ca. 2–4-fold enhanced potency of inhibitor 35 compared to its d-hPhe analog 36 for all enzymes.
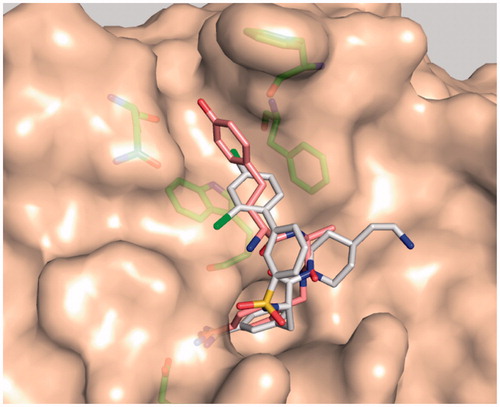
Based on the approximately 50-fold improved potency of inhibitor MI-432 () compared to its analog lacking the two chlorine atoms (Ki = 110 nM)Citation54, we expected a beneficial effect after incorporation of chloro-substituted d-hPhe analogs. However, all of these analogs, including the p-methoxy-derivatives 27 and 41, provided less potent matriptase inhibitors. A superimposition of inhibitor 35 and MI-432 reveals that the d-hTyr phenyl ring penetrates much deeper into the S3/4 pocket. It occupies a slightly different position compared to the dichloro-substituted phenyl ring of the 3-amidinophenylalanine-derived inhibitor, which makes a closer contact to the phenyl ring of Phe99 (). Moreover, the d-hTyr ring is more vertically oriented against Trp215, whereas the terminal dichloro-substituted phenyl ring of inhibitor MI-432 adopts a rather parallel orientation above the indole and vertically points against Phe99. These differences may explain why we could not achieve any improvement after incorporation of chloro- or methoxy-substituted d-hPhe analogs. So far, d-hTyr is the preferred residue in P3 position of this matriptase inhibitor type.
Figure 5. Model of matriptase in complex with inhibitor MI-432 (sticks with white carbon atoms, oxygen in red, nitrogen in blue, chlorine in green, structure shown in ) and the d-hTyr-derivative 35 (sticks with light pink carbons). The superpositions of both inhibitors reveals that their terminal phenyl rings occupy different positions in the distal S3/4 binding pocket. The matriptase residues Phe97, Phe99, Gln175, and Trp215, which shape the S3/4 pocket, and Asp189 are shown as sticks with green carbon atoms.
Conclusion
Starting from highly potent benzylsulfonyl protected substrate analog inhibitors of thrombin and fXa we could modulate their selectivity profile and obtained inhibitors with superior matriptase affinity. This was achieved by a combination of three modifications: (i) the elimination of the P4 benzylsulfonyl group, (ii) incorporation of d-hPhe analogs in P3 position, and (iii) replacement of the P3 Pro by Ala. However, we have to admit that the inhibitors 35 and 36 are still stronger trypsin inhibitors, although trypsin is mainly located in the intestinal tract and should not come in direct contact with injectable inhibitors. Presently, we cannot exclude that these compounds may also bind to other trypsin-like serine proteases, which were not tested so far. Moreover, the presence of two basic groups at the P3 and P1 residues makes it very unlikely that the unprotected inhibitors lacking the P4 moiety could be orally available. Nevertheless, the new inhibitors 35 and 36 are easily accessible and stable compounds and may serve as suitable tools for further biochemical studies with matriptase.
Declaration of interest
The authors report no conflicts of interest.
The authors acknowledge a travel grant from the Helmholtz Zentrum Berlin.
Supplementary material available online
IENZ_1172574_Supplementary_Material.pdf
Download PDF (142.1 KB)Acknowledgements
The authors thank the beamline support staff at BESSY for their advice during data collection.
References
- Antalis TM, Bugge TH, Wu Q. Membrane-anchored serine proteases in health and disease. Prog Mol Biol Transl Sci 2011;99:1–50
- Miller GS, List K. The matriptase-prostasin proteolytic cascade in epithelial development and pathology. Cell Tissue Res 2013;351:245–53
- List K, Haudenschild CC, Szabo R, et al. Matriptase/MT-SP1 is required for postnatal survival, epidermal barrier function, hair follicle development, and thymic homeostasis. Oncogene 2002;21:3765–79
- Basel-Vanagaite L, Attia R, Ishida-Yamamoto A, et al. Autosomal recessive ichthyosis with hypotrichosis caused by a mutation in ST14, encoding type II transmembrane serine protease matriptase. Am J Hum Genet 2007;80:467–77
- List K, Currie B, Scharschmidt TC, et al. Autosomal ichthyosis with hypotrichosis syndrome displays low matriptase proteolytic activity and is phenocopied in ST14 hypomorphic mice. J Biol Chem 2007;282:36714–23
- Buzza MS, Netzel-Arnett S, Shea-Donohue T, et al. Membrane-anchored serine protease matriptase regulates epithelial barrier formation and permeability in the intestine. Proc Natl Acad Sci USA 2010;107:4200–5
- Netzel-Arnett S, Buzza MS, Shea-Donohue T, et al. Matriptase protects against experimental colitis and promotes intestinal barrier recovery. Inflamm Bowel Dis 2012;18:1303–14
- Uhland K. Matriptase and its putative role in cancer. Cell Mol Life Sci 2006;63:2968–78
- Takeuchi T, Harris JL, Huang W, et al. Cellular localization of membrane-type serine protease 1 and identification of protease-activated receptor-2 and single-chain urokinase-type plasminogen activator as substrates. J Biol Chem 2000;275:26333–42
- Lee SL, Dickson RB, Lin CY. Activation of hepatocyte growth factor and urokinase/plasminogen activator by matriptase, an epithelial membrane serine protease. J Biol Chem 2000;275:36720–5
- Suzuki M, Kobayashi H, Kanayama N, et al. Inhibition of tumor invasion by genomic down-regulation of matriptase through suppression of activation of receptor-bound pro-urokinase. J Biol Chem 2004;279:14899–908
- Comoglio PM, Giordano S, Trusolino L. Drug development of MET inhibitors: targeting oncogene addiction and expedience. Nat Rev Drug Discov 2008;7:504–16
- Trusolino L, Comoglio PM. Scatter-factor and semaphorin receptors: cell signalling for invasive growth. Nat Rev Cancer 2002;2:289–300
- Shi X, Gangadharan B, Brass LF, et al. Protease-activated receptors (PAR1 and PAR2) contribute to tumor cell motility and metastasis. Mol Cancer Res 2004;2:395–402
- Milner JM, Patel A, Davidson RK, et al. Matriptase is a novel initiator of cartilage matrix degradation in osteoarthritis. Arthritis Rheum 2010;62:1955–66
- Chen CJ, Wu BY, Tsao PI, et al. Increased matriptase zymogen activation in inflammatory skin disorders. Am J Physiol Cell Physiol 2011;300:C406–15
- Hamilton BS, Gludish DW, Whittaker GR. Cleavage activation of the human-adapted influenza virus subtypes by matriptase reveals both subtype and strain specificities. J Virol 2012;86:10579–86
- Baron J, Tarnow C, Mayoli-Nüssle D, et al. Matriptase, HAT, and TMPRSS2 activate the hemagglutinin of H9N2 influenza A viruses. J Virol 2013;87:1811–20
- Galkin AV, Mullen L, Fox WD, et al. CVS-3983, a selective matriptase inhibitor, suppresses the growth of androgen independent prostate tumor xenografts. Prostate 2004;61:228–35
- Steinmetzer T, Schweinitz A, Stürzebecher A, et al. Secondary amides of sulfonylated 3-amidinophenylalanine. New potent and selective inhibitors of matriptase. J Med Chem 2006;49:4116–26
- Uhland K, Siphos B, Arkona C, et al. Use of IHC and newly designed matriptase inhibitors to elucidate the role of matriptase in pancreatic ductal adenocarcinoma. Int J Oncol 2009;35:347–57
- Zoratti GL, Tanabe LM, Varela FA, et al. Targeting matriptase in breast cancer abrogates tumour progression via impairment of stromal-epithelial growth factor signalling. Nat Commun 2015;6:6776
- Colombo E, Desilets A, Duchene D, et al. Design and synthesis of potent, selective inhibitors of matriptase. ACS Med Chem Lett 2012;3:530–4
- Han Z, Harris PK, Jones DE, et al. Inhibitors of HGFA, matriptase, and hepsin serine proteases: a nonkinase strategy to block cell signaling in cancer. ACS Med Chem Lett 2014;5:1219–24
- Goswami R, Mukherjee S, Wohlfahrt G, et al. Discovery of pyridyl bis(oxy)dibenzimidamide derivatives as selective matriptase inhibitors. ACS Med Chem Lett 2013;4:1152–7
- Goswami R, Wohlfahrt G, Mukherjee S, et al. Discovery of O-(3-carbamimidoylphenyl)-l-serine amides as matriptase inhibitors using a fragment-linking approach. Bioorg Med Chem Lett 2015;25:616–20
- Sisay MT, Steinmetzer T, Stirnberg M, et al. Identification of the first low-molecular-weight inhibitors of matriptase-2. J Med Chem 2010;53:5523–35
- Lila C, Gloanec P, Cadet L, et al. Large scale preparation of protected 4-aminomethylbenzamidine. application to the synthesis of the thrombin inhibitor, Melagatran. Chem Commun 1997;28:4419–29
- Gustafsson D, Bylund R, Antonsson T, et al. A new oral anticoagulant: the 50-year challenge. Nat Rev Drug Discov 2004;3:649–59
- Schweinitz A, Steinmetzer T, Banke IJ, et al. Design of novel and selective inhibitors of urokinase-type plasminogen activator with improved pharmacokinetic properties for use as antimetastatic agents. J Biol Chem 2004;279:33613–22
- Stürzebecher A, Dönnecke D, Schweinitz A, et al. Highly potent and selective substrate analogue factor Xa inhibitors containing D-homophenylalanine analogues as P3 residue: part 2. ChemMedChem 2007;2:1043–53
- Kadono S, Sakamoto A, Kikuchi Y, et al. Structure-based design of P3 moieties in the peptide mimetic factor VIIa inhibitor. Biochem Biophys Res Commun 2005;327:589–96
- Saupe SM, Steinmetzer T. A new strategy for the development of highly potent and selective plasmin inhibitors. J Med Chem 2012;55:1171–80
- Hellstern P, Stürzebecher U, Wuchold B, et al. Preservation of in vitro function of platelets stored in the presence of a synthetic dual inhibitor of factor Xa and thrombin. J Thromb Haemost 2007;5:2119–26
- Becker GL, Sielaff F, Than ME, et al. Potent inhibitors of furin and furin-like proprotein convertases containing decarboxylated P1 arginine mimetics. J Med Chem 2010;53:1067–75
- Kauer J. P-Benzoyl-L-phenylalanine, a new photoreactive amino acid. Photolabeling of calmodulin with a synthetic calmodulin-binding peptide. J Biol Chem 1986;261:10695–700
- Meyer D, Sielaff F, Hammami M, et al. Identification of the first synthetic inhibitors of the type II transmembrane serine protease TMPRSS2 suitable for inhibition of influenza virus activation. Biochem J 2013;452:331–43
- Walsmann P. On the purification of thrombin preparations. Pharmazie 1968;23:401–2
- Hammami M, Rühmann E, Maurer E, et al. New 3-amidinophenylalanine-derived inhibitors of matriptase. MedChemComm 2012;3:807–13
- Dixon M. The determination of enzyme inhibitor constants. Biochem J 1953;55:170–1
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol 1997;276:307–26
- Brandt T, Holzmann N, Muley L, et al. Congeneric but still distinct: how closely related trypsin ligands exhibit different thermodynamic and structural properties. J Mol Biol 2011;405:1170–87
- Collaborative Computational Project Number 4.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 1994;50:760–3
- Adams PD, Afonine PV, Bunkoczi G, et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010;66:213–21
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004;60:2126–32
- Bode W, Mayr I, Baumann U, et al. The refined 1.9 A crystal structure of human alpha-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment. EMBO J 1989;8:3467–75
- Tziridis A, Rauh D, Neumann P, et al. Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues. Biol Chem 2014;395:891–903
- Chemical Computing Group. MOE, version 2014.9, 32 bit. Montreal, Canada
- Sielaff F, Böttcher-Friebertshäuser E, Meyer D, et al. Development of substrate analogue inhibitors for the human airway trypsin-like protease HAT. Bioorg Med Chem Lett 2011;21:4860–4
- Biela A, Sielaff F, Terwesten F, et al. Ligand binding stepwise disrupts water network in thrombin: enthalpic and entropic changes reveal classical hydrophobic effect. J Med Chem 2012;55:6094–110
- Böttcher-Friebertshäuser E, Freuer C, Sielaff F, et al. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J Virol 2010;84:5605–14
- Schweinitz A, Stürzebecher A, Stürzebecher U, et al. New substrate analogue inhibitors of factor Xa containing 4-amidinobenzylamide as P1 residue: part 1. Med Chem 2006;2:349–61
- Tucker TJ, Lumma WC, Mulichak AM, et al. Design of highly potent noncovalent thrombin inhibitors that utilize a novel lipophilic binding pocket in the thrombin active site. J Med Chem 1997;40:830–2
- Steinmetzer T, Dönnecke D, Korsonewski M, et al. Modification of the N-terminal sulfonyl residue in 3-amidinophenylalanine-based matriptase inhibitors. Bioorg Med Chem Lett 2009;19:67–73