Abstract
Environmental exposures and genetic susceptibility can contribute to lung function decline in chronic obstructive pulmonary disease (COPD). Cigarette smoking is the main etiological factor for decline in lung function in COPD. However, only 10–20% chronic smokers develop symptomatic COPD. Genetic susceptibility to COPD might depend upon the variation of enzyme activities that detoxify cigarette smoke components. We performed a case control study to assess the association of Glutathione- S-transferase T1(GSTT1),Glutathione- S-transferase M1 (GSTM1), and Glutathione-S-transferase M3(GSTM3) common polymorphisms with the susceptibility to COPD patient in a north India population. In the present study, the genotypes of 412 subjects, (204 COPD patients and 208 healthy controls) were analyzed. Statistical analysis revealed that the frequency of homozygous GSTM1 null genotype was found to be significant higher in COPD patients as compared with healthy controls (OR, 2.58; 95% CI, 1.73–3.84; P = 0.001), but there were no significant differences in the distribution of homozygous null GSTT1 and 3-bp deletion polymorphism (rs1799735) in intron 6 variant allele in GSTM3 between COPD patients and healthy controls. Our study results suggest that GSTM1 null polymorphism is associated with genetic susceptibility to COPD. Moreover, we also found association between this polymorphism with pulmonary function test in smokers as well as nonsmokers.
INTRODUCTION
Chronic obstructive pulmonary disease (COPD) represents a major public health care problem worldwide due to its increasing prevalence, morbidity, and mortality (Citation1). Cigarette smoking is the most important risk factor for COPD, 10–20% of all cigarette smokers develop COPD (Citation2, 3). In addition to smoking, other environmental and genetic factors and gene environment interactions influence the development of COPD (Citation4), suggesting that individual susceptibility or genetic factors may play a role (Citation5).
The enzymes that detoxify constituents of tobacco smoke or its metabolites (Citation3) are good candidates for the study of inter-individual susceptibility to COPD. These enzymes include microsomal epioxide hydolase (mEPHX), glutathione-S-transferases (GSTs), and cytochrome P4501A1 (CYP1A1) (Citation6). Glutathione-S-transferases (GSTs) play inportant role in the detoxification of carcinogenic compounds contained in cigarette smoke and also in the antioxidant protection (Citation7, 8). The null genotypes of GSTM1 and GSTT1 genes affect the GSTmu and GSTtheta enzyme activities. The GSTmu has been recognized to detoxify smoke-derived carcinogens, e.g., polycyclic aromatic hydrocarbon and aromatic amines (Citation9). The homozygous GSTM1-null genotype has also been reported to show some association with the susceptibility of lung cancer (Citation10), emphysema (Citation11), and COPD (Citation12, 13).
GSTtheta conjugates glutathione and various potential carcinogens including halomethanes, (Citation14) which are present in cigarette smoke, and its null type mutant allele has been suggested as a risk factors for various diseases (Citation6,Citation15). Hu et al. found GSTT1 null genotype to be significantly associated with COPD in the Chinese population (Citation16, 17, Citation18), but other studies showed no association of null GSTT1 in COPD patients (Citation13 Citation19, 20, Citation21).
GSTM3 plays a role in the metabolism of harmful agents, like polyaromatic hydrocarbons benzo(α)pyrene, and has overlapping substrate specificity with GSTM1 (Citation13). GSTM3 gene, 3-bp deletion polymorphism (rs1799735) in intron 6 has been described as GSTM3A wild-type and GSTM3B variant alleles. The GSTM3B variant allele contains a recognition motif for the YY1 transcription factor, which has been postulated to regulate gene expression (Citation22). The GSTM3 polymorphism could, therefore, confer different efficiencies in the metabolism of tobacco smoke. GSTM3AA genotype has been reported to be associated with increased risk of larynx cancer while GSTM3AB genotype confers high risk for cervix cancer (Citation23) and GSTM3 AB+BB genotype were risk for gastric and esophageal cancers (Citation24, 25).
When multiple genes operate in the pathogenesis of disease, influence of each gene might be relatively weak (Citation26). Multiple genetic polymorphisms should be investigated to clarify whether the genetic events have additive effects or can predict the risk of developing COPD. Some previous studies have examined the role of GSTT1 and GSTM1 polymorphisms in COPD but no study has been carried out for GSTM3 polymorphism. Here, we have aimed to analyze the association GSTM1, GSTM1, and GSTM3 gene polymorphism with COPD patients in a cohort from a North Indian population, which is under-studied in terms of genetic association studies in COPD.
MATERIAL AND METHODS
Patients of COPD were diagnosis according to Global Initiative for chronic obstructive lung disease (GOLD) (Citation1, 2) were recruited from the outpatient Department of Pulmonary Medicine, Chhatrapati Shahuji Maharaj Medical University Lucknow, CSMMU (Erstwhile King George's Medical College). A total of 412 subjects (204 COPD patients and 208 healthy controls) were enrolled after evaluation of lung function by spirometry.
The COPD group consisted of 179 males and 25 females (mean age 57.83 ± 10.58 years) and healthy control group had 166 males and 42 females (mean age 56.35 ± 8.12 years). Exclusion criteria were respiratory disorders other than COPD, such as interstitial lung disease, any type of malignancy, previous history of asthma, tuberculosis. Healthy age-matched subjects without pervious medical history of respiratory disorders and without any present respiratory symptoms (Persistent cough, sputum production, or dyspnoea), seen by their practitioner for regular checkup, were recruited consecutively as controls. The participants were residence of geographic area of north India. The study was approved by ethical committee of CSMMU (Approval Code No-XXVIII ECM-B/P3), and the subjects were enrolled in accordance with the ethical standards of the Helsinki declaration of 1975. All subjects gave their written informed consent.
Spirometry
In all the subjects, pulmonary function tests were performed using Spiro 230 system (Morgan Scientific Inc, USA) 20 minutes before and after 200 μg dose of inhaled salbutamol (Foracort, CIPLA). Short-acting inhaled drugs were not used 6 hours before the testing, and long acting bronchodilator therapy was stopped for 12 hours before the test. All pulmonary function tests were performed according to the GOLD (Citation2) with the patients in the sitting position. FEV1 (forced expiratory volume in Citation1 second) adjusted for age, height, and sex. the inclusion criteria predicted post bronchodilator FEV1 <80%, FEV1/ forced vital capacity (FVC) <70% and improvement in FEV1 <12% or 200 ml after inhalation of 200 μg salbutamol of pre-bronchodilator (Citation2).
Blood collection and DNA extraction
EDTA-buffered whole blood (Citation5 ml) was drawn for subsequent DNA extraction by standard salting-out method (Citation27).
Genotype analysis
Null alleles ofGSTM1 and GSTT1 were determined by using multiplex polymerase chain reaction (PCR) with the CYP1A1 gene as an internal positive control (Citation28). Briefly, a 215-bp region between exons 4 and 5 of the GSTM1 gene and 480-bp products for were amplified along with a 312-bp size product of CYP1A1. The PCR products were electrophoresed on a 2% agarose gel. The absence of 480 and 215 bp bands indicated homozygous null genotypes of GSTM1 and GSTT1, respectively.
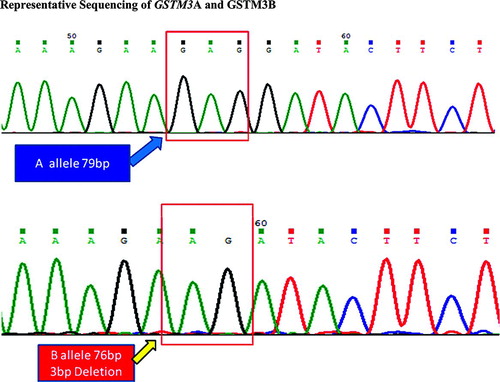
A simple PCR strategy was used to identify A and B alleles of the GSTM3 gene. Primers for intron 6 of GSTM3 were used to amplify a 79/76 bp product and electrophoretic separation on 20% polyacrylamide gel (Citation25). All reactions were carried out in an MJ Research 200 thermal cycler. PCR products and molecular weight markers were visualized after staining with ethidium bromide (Etbr).
DNA sequencing
PCR amplicons of GSTM3 were purified and sequenced using automated DNA sequencer. The sequence analysis was performed using DNA STAR and Chromas 231 (available online) software. .
Statistical analysis
Descriptive statistics of patients and controls were presented as mean and standard deviations for continuous measures whereas frequencies and percentages were used for categorical measures. Effective sample sizes were calculated by the Quanto software version 1.2(Gauderman WJ, Morrison JM. QUANTO 1.1 a computer program for power and sample size calculations for genetic-epidemiology studies <http://hydra.usc.edu/gxe> (2006). Independent sample t-test was performed to compare the mean values of continuous data and χ2-test use for to compare the genotype frequencies between patient and control. The clinical parameters were expressed as Mean ± SD. Odds ratio (OR), 95% confidence interval (CI) and p value of genotype data were adjusted for confounding factors such as age, sex and pack-years of smoking. A p value of ≤0.05 was considered statistically significant.
RESULTS
Patients and Controls
Twenty-five of the 179 COPD patients and 42 of the 166 healthy controls were female, while the rest were male (). No significant differences were observed in age (p = 0.111). Numbers of smokers were higher in patient's comparison to healthy controls. Significant difference was found in pack-year smoked (p = 0.001), in control was 13.51 ± 6.97 pack-years. But patients with COPD had a 23.67 ± 17.89 pack-year's history of smoking. In COPD patient significant lower lung function was observed as compared with the healthy control, their pulmonary function tests showed predicted mean post-bronchodilator FEV1 of 40.34 ± 12.96%, and predicted (FVC) of 53.31 ± 12.90%, with mean FEV1/FVC ratio 54.85 ± 14.22%, while healthy control showed FEV1 of 80.82 ± 23.36%, FVC 84.75 ± 19.80%, with mean FEV1/FVC ratio 94.54 ± 14.04%.
Table 1. Demographic characteristic of COPD cases and controls according to GOLD criteriaFootnote*
Genotypes of GSTM1 and GSTT1
The distribution of genotypes and allele frequencies of GSTM1 and GSTT1 in patients and controls and the respective ORs are presented in . The frequency of GSTM1 null genotypes was higher (60.8 vs. 37.5%) and imposed significant 2-fold risk for COPD patients (OR, 2.58; 95% CI, 1.73–3.84; P = 0.001). It was observed that when FEV1/FVC less than 70% was significantly associated with GSTM1 null genotype, data was not showed. However, the frequency of GSTT1 null genotypes were not found to be significantly different between COPD patients and healthy control (17.6 vs 24.5%,).
Table 2. Frequency distribution of GSTM1, GSTT1 and GSTM3 between COPD cases and control
Genotype of GSTM3
In controls, observed genotype frequency of GSTM3 polymorphism did not deviate from Hardy-Weinberg equilibrium The frequency of AB genotype of GSTM3 was higher in COPD patient in comparison to healthy control (34.3% vs. 28.8%), but difference was non-significant (OR, 1.75; 95% CI, 0.28–10.32; P = 0.547) ().
Gene-environment interaction
shows the association between the different genotypes with COPD in smokers and nonsmokers. The GSTM1 null polymorphism was associated with an increased risk of developing COPD in both the smokers (OR, 4.05; 95% CI, 1.99–8.27; P = 0.001), as well as non-smokers (OR, 2.78; 95% CI, 1.47–5.27; P = 0.002), while no differences were observed for GSTT1 and GSTM3 polymorphisms.
Table 3. Association of GSTT1, GSTM1 and GSTM3 polymorphism with COPD cases and control on basis of cigarette smoking
DISCUSSION
There is likely to be a complex interplay between environmental and genetic factors for the development of COPD. It has been suggested that individual susceptibility due to genetic predisposition is likely to be an important determinant of this disease. Genetic polymorphisms in xenobiotic enzyme genes may play a role in susceptibility to oxidant related lung disease, and several studies have assessed the association between COPD and candidate genes of enzymes involved in the metabolism of mutagens and carcinogens originating in cigarette smoke components that are believed to cause some of the lung damage characteristic of COPD (Citation29).
Glutathione S-transferases (GSTs) are a large family of enzymes participating in detoxification of endogenous and exogenous toxic substrates including tobacco derived toxins; they also exhibit peroxidase activity and thus might play an important role in oxidative stress (Citation30).
Our results show clear association of null genotype of GSTM1 with COPD while GSTT1 null or GSTM3 intron 6 polymorphism do not play role in the disease susceptibility, In case of COPD patients, the frequency of GSTM1 null genotype was higher in cases compared to controls (OR, 2.58; 95% CI, 1.73–3.84; P = 0.001). The results in GSTM1 are agreement with previous studies on COPD (Citation15, Citation31). The null allele of GSTM1 may result in defective detoxification of the polycyclic aromatic hydrocarbons of cigarette smoke and may promotes cellular and tissue damage of the lung due to an excess of oxidants and free radicals (Citation32).
The frequency of GSTM1-null genotype in the control group was closer to that found in Western countries as well as other Indian reports (37.5 versus 46.9–53%),but it was lower than of Koreans population (65%) (Citation4, Citation11, Citation25, Citation33). The frequency of GSTT1-null genotype shows distinct difference in various ethnic groups, it is 10–18%, in Caucasian population and 22–29% in African Americans (Citation34). In the present study, frequency of controls group (24.5%) was similar to other north India populations but lower than various Asian populations (Citation19), However, no association of null GSTT1 with COPD (Citation12, Citation19, Citation21) is in conformity with several previous studies (Citation12, 13, Citation19, Citation21).
GSTM3 3bp deletion intron 6 has two alleles: GSTM3A and GSTM3B, The 3bp deletion generates a site for the binding of transcription factor YY1 which can both repress and activate transcription. However, molecular mechanisms controlling its behavior are complex and still not understood (Citation6), The frequency of GSTM3 B mutant allele has been rated as 15% to 24% (Citation35) in Caucasians and 68% in African Americans (Citation35). In the present study, frequency of GSTM3 B allele was 15.9% in controls, which is closer to Caucasians but different from other Indian studies (Citation1, Citation25, Citation36). To the best of our knowledge, this is the first study to examine association of GSTM3B mutant allele in COPD but genotype and allelic frequencies of GSTM3 polymorphic alleles were similar in COPD and control groups.
In our study we analyzed the association of genotypes of the GSTT1, GSTM1, and GSTM3 genes with the smoking history and pulmonary function test (post-bronchodilator FEV1/FVC%). We observed significant association of GSTM1 null in both smokers and non-smokers. Although cigarette smoking is major risk factor for COPD but, there is a substantial body of epidemiological evidence linking occupational exposures to dusts, gases/vapors, and fumes to chronic airflow destruction, with a substantial population attributable risk (15–20%) in non-smokers (Citation37). In India 5% male and 2.7% females subjects are affected by COPD (Citation38), and tobacco smoking is common in most male patients. but non-smoker patients (especially women) are significantly exposed of solid fuels or occupational exposures to dust and fumes (Citation39). The smoke from combustion of solid fuels such as dried dung, wood and crop residue used for cooking and heating, especially in villages, semi-urban and slum areas, is an important cause of pollution of the indoor air, and it is responsible for large number of COPD patients in the rural inhalation in general and women in particular (Citation39).
Pillai et al. (Citation26) conducted a genome wide association study (GWAS) for gene environment interaction in COPD in a cohort of Norwegian samples and observed strong association with SNPs belonging to a-nicotinic acetylcholine receptor (CHRNA 3/5) locus. The absence of genes belonging to GST family in the GWAS may be related to population specific variations in candidate gene approaches.
The limitation of this study is small sample sizes and the fact that only a few genes involved in the detoxification of smoke products were studied. In conclusion, we found association of GSTM1 null polymorphism with COPD in a north India population. Moreover, we also found association between this polymorphism with pulmonary function test in smokers as well as nonsmokers with COPD. However, COPD is a multifactorial disease and role of other genes and their interactions should also be considered before their clinical applications.
ACKNOWLEDGMENT
The study was supported by a research grant (UPCST/SERPD-D-3404).from Council of Science & Technology, U.P., Lucknow.
Conflict of interest
None of the authors have conflict of interest to declare in relation to this work. The authors alone are responsible for the content and writing of the paper.
REFERENCES
- Celli BR, MacNee W. Standards for the diagnosis and treatment of patients with COPD: A summary of the ATS/ERS position paper. Eur Respir J 2004; 23:932–946.
- Global Initiative for Chronic Obstructive Lung Disease (GOLD). Global strategy for the diagnosis, Management and prevention of chronic obstructive pulmonary disease. Updated 2009. http://www.goldcopd.com/ Accessed October 2010.
- Sherrill DL, Lebowitz MD, Burrows B. Epidemiology of chronic obstructive pulmonary disease. Clin Chest Med 1990; 11:375–387.
- Sandford AJ, Silverman EK. Chronic obstructive pulmonary disease. 1: Susceptibility factors for COPD the genotype-environment interaction. Thorax 2002; 57:736–741.
- Laurell CB, Eriksson S. The electrophoretic alpha 1-globulin pattern of serum in alpha 1-antitrypsin deficiency. Scand J Clin Lab Invest 1963; 15:132–140.
- Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta 1997; 1332:F49–66.
- Ketterer B, Harris JM, Talaska G, Meyer DJ, Pemble SE, Taylor JB, The human glutathione S-transferase supergene family, its polymorphism, and its effects on susceptibility to lung cancer. Environ Health Perspect 1992; 98:87–94.
- He JQ, Ruan J, Connett JE, Anthonisen NR, Pare PD, Sandford AJ. Antioxidant gene polymorphisms and susceptibility to a rapid decline in lung function in smokers. Am J Respir Crit Care Med 2002; 166:323–328.
- Hirvonen A, Nylund L, Kociba P, Husgafvel-Pursiainen K, Vainio H. Modulation of urinary mutagenicity by genetically determined carcinogen metabolism in smokers. Carcinogenesis 1994; 15:813–815.
- Hirvonen A, Husgafvel-Pursiainen K, Anttila S, Vainio H. The GSTM1 null genotype as a potential risk modifier for squamous cell carcinoma of the lung. Carcinogenesis 1993; 14:1479–1481.
- Harrison DJ, Cantlay AM, Rae F, Lamb D, Smith CA. Frequency of glutathione S-transferase M1 deletion in smokers with emphysema and lung cancer. Hum Exp Toxicol 1997; 16:356–360.
- Zidzik J, Slaba E, Joppa P, Kluchova Z, Dorkova Z, Skyba P, Glutathione S-transferase and microsomal epoxide hydrolase gene polymorphisms and risk of chronic obstructive pulmonary disease in Slovak population. Croat Med J 2008; 49:182–191.
- Faramawy MM, Mohammed TO, Hossaini AM, Kashem RA, Abu Rahma RM. Genetic polymorphism of GSTT1 and GSTM1 and susceptibility to chronic obstructive pulmonary disease (COPD). J Crit Care 2009; 24:e7–10.
- Pemble S SK, Spencer SR, Meyer DJ, Hallier E, Bolt HM, Ketterer B, Taylor JB. Human glutathione S-transferase theta (GSTT1): cDNA cloning and the characterization of a genetic polymorphism. Biochem J 1994; 300:271–276.
- Chenevix-Trench G, Young J, Coggan M, Board P. Glutathione S-transferase M1 and T1 polymorphisms: Susceptibility to colon cancer and age of onset. Carcinogenesis 1995; 16:1655–1657.
- Hu G, Yao W, Zhou Y, Hu J, Shi Z, Li B, Meta- and pooled analyses of the effect of glutathione S-transferase M1 and T1 deficiency on chronic obstructive pulmonary disease. Int J Tuberc Lung Dis 2008; 12:1474–1481.
- Faramawy MM, Hossaini AM, Kashem RA, Abu Rahma RM. Genetic polymorphism of GSTT1 and GSTM1 and susceptibility to chronic obstructive pulmonary disease (COPD). J Crit Care 2009; 24:e7–e10.
- Mehrotra S, Sharma A, Kumar S, Kar P, Sardana S, Sharma JK. Polymorphism of glutathione S-transferase M1 and T1 gene loci in COPD. Int J Immunogenet 2010; 37:263–267.
- Chan-Yeung M, Ho SP, Cheung AH, So LK, Wong PC, Chan KK, Polymorphisms of glutathione S-transferase genes and functional activity in smokers with or without COPD. Int J Tuberc Lung Dis 2007; 11:508–514.
- Cheng SL, Yu CJ, Chen CJ, Yang PC. Genetic polymorphism of epoxide hydrolase and glutathione S-transferase in COPD. Eur Respir J 2004; 23:818–824.
- Yim JJ, Park GY, Lee CT, Kim YW, Han SK, Shim YS, Genetic susceptibility to chronic obstructive pulmonary disease in Koreans: Combined analysis of polymorphic genotypes for microsomal epoxide hydrolase and glutathione S-transferase M1 and T1. Thorax 2000; 55: 121–125.
- Inskip A, Elexperu-Camiruaga J, Buxton N, Dias PS, MacIntosh J, Campbell D, Identification of polymorphism at the glutathione S-transferase, GSTM3 locus: Evidence for linkage with GSTM1*A. Biochem J 1995; 312 (Pt 3):713–716.
- Singh H, Sachan R, Devi S, Pandey SN, Mittal B. Association of GSTM1, GSTT1, and GSTM3 gene polymorphisms and susceptibility to cervical cancer in a North Indian population. Am J Obstet Gynecol 2008; 198:303e1–6.
- Malik MA, Upadhyay R, Mittal RD, Zargar SA, Modi DR, Mittal B. Role of xenobiotic-metabolizing enzyme gene polymorphisms and interactions with environmental factors in susceptibility to gastric cancer in Kashmir Valley. J Gastrointest Cancer 2009; 40:26–32.
- Jain M, Kumar S, Lal P, Tiwari A, Ghoshal UC, Mittal B. Role of GSTM3 polymorphism in the risk of developing esophageal cancer. Cancer Epidemiol Biomarkers Prev 2007; 16:178–181.
- Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, A genome-wide association study in chronic obstructive pulmonary disease (COPD): Identification of two major susceptibility loci. PLoS Genet 2009; 5:e1000421.
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16:1215.
- Setiawan VW, Zhang ZF, Yu GP, Li YL, Lu ML, Tsai CJ, GSTT1 and GSTM1 null genotypes and the risk of gastric cancer: a case-control study in a Chinese population. Cancer Epidemiol Biomarkers Prev 2000; 9:73–80.
- Smith CA, Harrison DJ. Association between polymorphism in gene for microsomal epoxide hydrolase and susceptibility to emphysema. Lancet 1997; 350:630–633.
- Hayes JD, Strange RC. Glutathione S-transferase polymorphisms and their biological consequences. Pharmacology 2000; 61:154–166.
- Baranova H, Perriot J, Albuisson E, Ivaschenko T, Baranov VS, Hemery B, Peculiarities of the GSTM1 0/0 genotype in French heavy smokers with various types of chronic bronchitis. Hum Genet 1997; 99:822–826.
- Imboden M, Downs SH, Senn O, Matyas G, Brandli O, Russi EW, Glutathione S-transferase genotypes modify lung function decline in the general population: SAPALDIA cohort study. Respir Res 2007; 8:2.
- Ryberg D, Skaug V, Hewer A, Phillips DH, Harries LW, Wolf CR, Genotypes of glutathione transferase M1 and P1 and their significance for lung DNA adduct levels and cancer risk. Carcinogenesis 1997; 18:1285–1289.
- Geisler SA, Olshan AF. GSTM1, GSTT1, and the risk of squamous cell carcinoma of the head and neck: a mini-HuGE review. Am J Epidemiol 2001; 154:95–105.
- Jourenkova-Mironova N, Voho A, Bouchardy C, Wikman H, Dayer P, Benhamou S, Glutathione S-transferase GSTM3 and GSTP1 genotypes and larynx cancer risk. Cancer Epidemiol Biomarkers Prev 1999; 8:185–188.
- Pandey SN, Jain M, Nigam P, Choudhuri G, Mittal B. Genetic polymorphisms in GSTM1, GSTT1, GSTP1, GSTM3 and the susceptibility to gallbladder cancer in North India. Biomarkers 2006; 11:250–261.
- Meldrum M, Rawbone R, Curran AD, Fishwick D. The role of occupation in the development of chronic obstructive pulmonary disease (COPD). Occup Environ Med 2005; 62: 212–214.
- Jindal SK, Aggarwal AN, Gupta D. A review of population studies from India to estimate national burden of chronic obstructive pulmonary disease and its association with smoking. Ind J Chest Dis Allied Sci 2001; 43:139–147.
- Jindal SK, Gupta D, Aggarwal AN. Guidelines for management of chronic obstructive pulmonary disease (COPD) in India: A guide for physicians (2003). Ind J Chest Dis All Sci 2004; 46:137–153