Abstract
Surfactant protein D (SFTPD) is a lung-specific anti-inflammatory factor that antagonizes inflammation by inhibiting oxidative stress and stimulating innate immunity. Variations in SFTPA2 and SFTPB, genes for other surfactant proteins, have been associated with lung cancer. We therefore investigated associations between SFTPD variations and lung cancer as well as emphysema and interstitial pneumonia, which are characterized by chronic inflammation from which lung cancer often arises. DNA from 1342 autopsy samples, including those from 140 subjects with lung cancer, was investigated. The single nucleotide polymorphism (SNP) rs721917, which results in methionine being exchanged for threonine at amino acid 11 (the Met11Thr variation), tended to be associated with emphysema and was associated with interstitial pneumonia and lung cancer. A haplotype analysis revealed that the haplotypes associated with emphysema and lung cancer differed from that associated with interstitial pneumonia, suggesting a differential role for SFTPD in the development of these diseases. A mediating analysis did not reveal a mediating effect exerted by emphysema or interstitial pneumonia on lung cancer. Our results suggested that SFTPD plays a role in the development of lung cancer and that the role for lung cancer may differ from that for interstitial pneumonia.
Introduction
Lung cancer is a leading cause of cancer-related death worldwide (Citation1, 2) and one of the most lethal forms of cancer in Japan (Citation3). Genetic susceptibility in the development of lung cancer has been suggested by familial clustering and by the results of segregation analysis of lung cancer (Citation4). Recent genome-wide association studies have suggested that variations in the genes comprising the nicotinic acetylcholine receptor gene cluster, which contains multiple genes affecting nicotine dependence of individuals, contribute to susceptibility to lung cancer in Caucasians (Citation5–7) as well as in Japanese (Citation8). However, whether these variations directly contribute to lung cancer or indirectly contribute to its development through smoking behavior remains controversial. Further, the genetic variations that directly affect the molecular mechanism of carcinogenesis remain largely unknown.
Chronic inflammation is a risk factor for cancer (Citation9). The carcinogenic effect of smoking is partly mediated by the induction of diseases characterized by chronic inflammation including emphysema (Citation10, 11) and interstitial pneumonia (IP) (Citation12), which frequently accompany lung cancer. A study has reported that chronic obstructive pulmonary disease (COPD) patients treated with inhaled corticosteroids have a reduced incidence of lung cancer, which suggests an important role of chronic inflammation in carcinogenesis (Citation13). Therefore, genetic variations that affect chronic inflammation may also influence the incidence of lung cancer.
Surfactant protein D (SFTPD) is a lung-specific anti-inflammatory factor that is expressed in alveolar type II cells. It antagonizes inflammation by inhibiting oxidative stress (Citation14) and stimulating innate immunity (Citation15). Genetic association studies have shown that variations in the genes SFTPA2 and SFTPB, which encode other surfactant proteins sharing several functions with SFTPD, are associated with lung cancer (Citation16, 17). Furthermore, SFTPD variations have been associated with bronchial dysplasia, which is a known precancerous condition (Citation18). These observations suggest that SFTPD may have a role in lung carcinogenesis.
We have previously identified an association between genetic variation in SFTPD and susceptibility to emphysema in Japanese patients (Citation19). In the current study, we extended the analysis and investigated the interrelationships between SFTPD and emphysema, IP, and lung cancer from a genetic standpoint. For this, we focused on the single nucleotide polymorphism (SNP) rs721917 in SFTPD, which results in an amino acid change from Met (“T” allele) to Thr (“C” allele) at amino acid 11, and the SNP rs10887199, which does not cause any amino acid alterations but has been associated with emphysema (Citation19, 20). We analyzed the associations of emphysema, IP, and lung cancer with these SNPs by using 1342 autopsy samples that included samples from 140 subjects with lung cancer.
Methods
Subjects
A total of 1536 patients were autopsied at the Department of Pathology, Tokyo Metropolitan Geriatric Medical Center, Tokyo, Japan, between February 1995 and July 2003. Of these, 1342 subjects who were of Japanese nationality and had a detailed pathological record of emphysema, IP, or lung cancer were enrolled in the study. The genomic DNA of each subject was used with written informed consent from a family member.
Pathological diagnosis of subjects
Each subject underwent pathological examination, and the emphysema type (centriacinar, panacinar, or paraseptal) and severity (0, none; 1, minimal; 2, mild; 3, moderate; or 4, severe) (Citation21, 22) was determined as described in our previous report (Citation23). Briefly, both excised lungs were infused with a 10% formalin solution at a transpulmonary pressure of 25 cm H2O. Sagittal slices approximately 2 cm thick were obtained. The lung slices were macroscopically assessed for emphysematous changes according to type and severity, and 3 pathologists (TA, TK, and MS) independently scored them without prior interpretation.
The scoring was confirmed to be devoid of inter- and intra-observer differences. The mean scores of the 3 pathologists for all slices of both lungs were used in the current study. Subjects with the centriacinar type with a severity of 3 or 4, and/or the panacinar type with a severity of >1, were included in the emphysema group. The main phenotypes of emphysema are centriacinar, panacinar, and paraseptal. We excluded the paraseptal type and only included the centriacinar and panacinar types because centriacinar emphysema is caused mainly by smoking and panacinar emphysema with centriacinar emphysema is an aggravated form of centriacinar emphysema (Citation24). Subjects who lacked centriacinar- or panacinar-type regions were included in the non-emphysema group.
The diagnosis of IP was based on the clinical and autopsy findings of TK, MS, and TA. The IP cases included idiopathic, acute, and collagen disease-associated IPs (Citation25, 26). Subjects with sarcoidosis and pneumoconiosis such as asbestosis were not included. We analyzed all clinical and pathological data to determine the presence or absence of lung cancer. We included cancers incidentally found at autopsy.
Genotyping
Genomic DNA was extracted from the renal cortex by using the phenol–chloroform method and stored at –20°C until use. The SNPs described above were genotyped using an ABI TaqMan® SNP genotyping assay (Life Technologies Japan, Tokyo, Japan) as previously described (Citation27).
Statistical analysis
All values are presented as mean (SD). The differences observed between groups were compared using the unpaired t-test, Wilcoxon test, or Pearson's |2 test. Statistical analyses were performed using JMP genomics software version 3.1 (SAS Institute Inc., Cary, NC, USA) or the Statistical Package for the Social Sciences (SPSS) for Windows version 11.0.1 (SPSS Inc., Chicago, IL, USA).
The effect of each genotype of rs721917 on emphysema, IP, or lung cancer was calculated using multivariate logistic regression with a dominant, recessive, or additive model based on the “T” allele (the minor allele). The adjusted odds ratio in each model was calculated as follows: In the dominant model, the ratio of the adjusted odds for the “T/T” plus “T/C” genotypes to that for the “C/C” genotype was evaluated. In the recessive model, the ratio of the adjusted odds for the “T/T” genotype to that for the “T/C” plus “C/C” genotypes was evaluated. In the additive model, the increment of the adjusted odds ratio per one “T” allele was evaluated. The effect of each genotype of rs10887199 was similarly evaluated on the basis of the “C” allele (the minor allele).
We estimated the unobserved haplotype frequencies by the expectation–maximization algorithm that assumes the Hardy–Weinberg equilibrium and then speculated the haplotype for each subject. The effect of each haplotype on emphysema, IP, or lung cancer was calculated using multivariate logistic regression with the additive model, in which age, gender, smoking status, and pack-years were used as covariates. The odds ratio (OR) for the development of emphysema, IP, or lung cancer according to the copy number of a chromosomal fragment with a specific haplotype was determined using a logistic regression model adjusted for age, sex, smoking status, and pack-years.
A mediation analysis was performed for the identification of a potential mediating effect that originates from the genetic variation in SFTPD and influences lung cancer through emphysema or IP (Citation28, 29). In the current study, the initial variable was the rs721917 genotype, the mediator was the presence of either emphysema or IP, and the outcome was lung cancer (Citation30, 31). First, a suitable model for the regression analysis was selected on the basis of the result of the association study, i.e., the additive model for emphysema and the recessive model for IP. In the additive model, the rs721917 genotype was coded as 0, 1, or 2 according to the number of the “T” allele. In the recessive model, the rs721917 genotype “T/T” was coded as 1 or 0. Second, the regression analysis adjusted for age, gender, smoking status, and pack-years was performed using 3 different conditions: (Citation1) the initial variable was an independent variable and the outcome was a dependent variable; (Citation2) the initial variable was an independent variable and the mediator was a dependent variable; and (Citation3) both the initial variable and the mediator were independent variables and the outcome was a dependent variable. Then, the mediating effect was calculated using the β parameter estimates obtained in each condition. The significance of the mediating effect was tested using the Sobel test (Citation30, Citation32). A p-value of <0.05 was considered significant.
Ethical considerations
The current study was approved by the ethics committees of Nippon Medical School (approval no. 18-11-31), Tokyo Medical and Dental University (approval no. 4), and Tokyo Metropolitan Geriatric Medical Center (approval no. 440).
Results
Subjects
The characteristics of the subjects are shown in The presence of emphysema was significantly associated with lung cancer. The lung-cancer group had a greater number of smokers, smokers with a greater number of pack-years, and a greater proportion of males than the group without lung cancer. We thus performed regression analyses with adjustments for age, gender, and pack-years. The prevalence of IP in this autopsy population was high. One of the reasons is thought to be a bias in the study population. Tokyo Metropolitan Geriatric Medical Center is a core hospital with a ward for respiratory medicine, and the proportion of IP is expected to be higher than that in the general population. Furthermore, the morbidity of idiopathic pulmonary fibrosis (IPF) is reported to be higher in aged populations (Citation33); therefore, the relatively high age of this population might also have contributed to the higher proportion of subjects with IP including IPF.
Table 1. Genotypes of SFTPD in the subjects with IP, emphysema, and lung cancer
SNP selection
We selected 2 SNPs (rs721917 and rs10887199) from the SFTPD gene region that have been associated with COPD (Citation19, 20) (Supplementary ). The SNP rs721917 (TC) is of great interest because it causes an amino acid change (Met11Thr) that results in a reduced serum concentration of the SFTPD protein product as well as a lower oligomerization ability and reduced binding to bacteria of SFTPD (Citation34, 35). The SNP rs10887199 is not located in the coding region but has been associated with COPD in our previous study (Citation19). These SNPs have a large minor allele frequency in the Japanese population (0.477 for the “T” allele of rs721917 and 0.344 for the “C” allele of rs10887199; HapMap database [http://www.hapmap.org]) and are thus suitable for use in an association study.
Association of SFTPD SNPs with emphysema, IP, and lung cancer
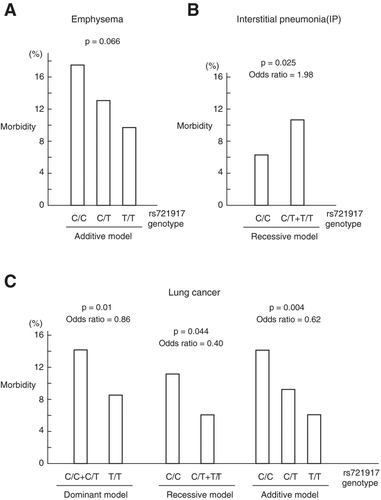
The genotypes of SFTPD found in subjects with emphysema, IP, and lung cancer are shown in Supplementary . We used the dominant, recessive, and additive models to investigate the effect of the SNP. In the dominant model, a single copy of the risk allele exerts a full effect. In the recessive model, 2 copies of the risk allele are required to produce a full effect, while a single copy does not have any effect. In the additive model, 2 copies of the risk allele are required for a full effect, while a single copy provides half of the full effect. Then, we investigated whether any of the models elucidated the association between each SNP and the development of emphysema, IP, or lung cancer. We found that rs721917 tended to be associated with emphysema (p = 0.066) in the additive model, although this association was not significant ().
Table 2. Associations of SFTPD with emphysema, IP, and lung cancer
We also found that rs721917 was associated with IP in the recessive model (p = 0.025) and with lung cancer in all models (p = 0.01–0.005), among which the additive model had the smallest p-value (). The rs10887199 SNP did not show any association in the current study. The risk allele of rs721917 for emphysema was probably the “C” allele (), for IP was the “T” allele (), and for lung cancer was the “C” allele (). These results suggest that the mechanism of involvement of SFTPD in emphysema may be the same as that in lung cancer, while the mechanism of SFTPD involvement in IP differs from that in lung cancer.
Association of SNP haplotypes at the rs721917/rs10887199 region with emphysema, IP, and lung cancer
In the analysis described here, the rs721917 “C” allele tended to be associated with emphysema and was associated with lung cancer, while the rs721917 “T” allele was associated with IP. To further confirm this result, we investigated the associations of their haplotypes with these diseases. As shown in Supplementary , rs721917 and rs10887199 belong to the same linkage disequilibrium block and are thus suitable for determining the local haplotype. The calculation indicated that the rs721917-C/rs10887199-C haplotype is associated with emphysema, while the rs721917-T/rs10887199-C haplotype is associated with IP (). This again indicated that the rs721917 genotype associated with emphysema differed from that associated with IP. Lung cancer was associated with both “T/T” and “C/T” haplotypes (). This indicates that the rs10887199 “T” genotype may also have an effect on lung cancer rather than the rs721917 genotype.
Table 3. Associations of the haplotype at the rs721917/rs10887199 region with emphysema, interstitial pneumonia, and lung cancer
Mediating effect analysis
In some cases, a factor exerts the final effect through an intermediate phenotype. In other words, a factor causes an intermediate phenotype, and the intermediate phenotype causes the final phenotype. The effect that a factor exerts on the final phenotype via an intermediate phenotype is called a mediating effect. At rs1051730 in the CHRNA5–3 region, COPD has been reported to be a mediator of lung cancer (Citation29). We tested our hypothesis that variations in SFTPD may be mediated by emphysema or IP, which are phenotypes characterized by chronic inflammation. A mediation analysis was employed to statistically test the presence of a mediating effect (Supplementary Figure 2).
We were unable to detect a statistically significant mediating effect of emphysema or IP (). This suggests that the mediating effect, if it exists at all, is not substantial.
Table 4. Mediation effect
Discussion
In the current study, we tested whether genetic variations in SFTPD are directly associated with lung cancer or indirectly contribute to lung cancer development through effects mediated by diseases that are characterized by chronic inflammation. The study employed 1342 consecutive autopsy subjects including 140 subjects with lung cancer. We observed a significant association of IP and lung cancer with a coding SNP that causes a Met11Thr amino acid change. The risk haplotypes of IP and lung cancer differed from that for emphysema. We were unable to detect a significant mediating effect of the SFTPD genotype on lung cancer through emphysema or IP.
The results of the association analyses for genotype and haplotype suggested that the role of SFTPD in the development of emphysema and lung cancer may differ from that in the development of IP.
Oxidative stress and bacterial infections are risk factors for emphysema (Citation14). The “T” allele of rs721917, which increases the serum concentration of SFTPD and produces SFTPD protein with a greater oligomerization ability and a greater ability to bind to bacteria (Citation34, 35), may prevent emphysema through an increased level of protein activity. Because the “C” allele of the rs721917 SNP was associated with both emphysema and cancer and the risk haplotypes for lung cancer differed from those for emphysema, a SNP other than rs721917, which is correlated not to “C/T” but to “C/C”, might also be associated with emphysema.
Two haplotypes in the SFTPD gene were associated with lung cancer in our study. The circulating level of SFTPD protein has recently been reported to be associated with subsequent lung cancer risk (Citation36), which also suggests an association between SFTPD and lung cancer. Although the haplotypes including the “T” allele of rs10887199 were associated with lung cancer, the SNP rs10887199 itself was not associated with this disease. One possible explanation is that these haplotypes are associated with lung cancer through different causal SNPs, which are not strongly correlated to rs10887199. Several groups suggested that different SNPs in one single gene are associated with diseases, e.g., cystic fibrosis, dyslipidemia, and lung cancer (Citation37–39).
The carbohydrate recognition domain of SFTPD protein binds deleted in malignant brain tumors 1 (DMBT1), a tumor suppressor, and the expression of DMBT1 is frequently lost in lung cancer (Citation40, 41). One of the possible mechanisms of the association of these SFTPD haplotypes and lung cancer is that these haplotypes may result in changes in the carbohydrate recognition domain, which affects the binding of SFTPD to DMBT1 and may reduce the tumor-suppressing capability of DMBT1.
Endoplasmic reticulum (ER) stress is possibly involved in the development of IP. A mutation in the SFTPC precursor protein has been reported to result in the accumulation of unfolded proteins in the ER, thereby increasing ER stress (Citation42, 43). The rs721917-T/rs10887199-C haplotype might tag a nucleotide change that causes a structural defect in the SFTPD protein.
There are some limitations in this study. First, we were unable to demonstrate a significant mediating effect of emphysema or IP on lung cancer; this may be due to the small sample size used in the current study. A study with a larger sample size may be needed for the detection of a weak mediating effect (Citation30). Second, we did not correct the results for multiple testing in this study. If Bonferroni corrections are performed, the thresholds of the p-value after correction for the numbers of SNPs, diseases, and genetic models, and for the numbers of both haplotypes and genetic models, are 0.05/(2 × 3 × 3) = 0.002 and 0.05/(3 × 3) = 0.005, respectively.
According to these strict thresholds, we cannot observe significant associations between the SNPs and the diseases in Tables and . An analysis of the association between genetic variations of these SNPs and the diseases by two-way analysis of variance with Tukey–Kramer's honestly significant difference test and with Bonferroni's adjustment for the number of the diseases provided a significant association between rs721917 and lung cancer. Another independent population would be needed to validate our results on the association between the genetic variations of SFTPD and emphysema, IP, and lung cancer.
Conclusions
In conclusion, genetic variation in SFTPD is a risk factor for the development of IP and lung cancer. Our observation that the genetic variation associated with emphysema and lung cancer differs from that associated with IP suggests differential roles for SFTPD in the development of these diseases.
Declaration of Interests
The authors have no conflicts of interest to disclose. A professional editing company, Editage, reviewed the manuscript for correct English usage. The authors are responsible for the content and the writing of this paper. This study was partly supported by a grant from the Grant-in-Aid for Young Scientists (B) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- Jemal A, Siegel R, Ward E, Cancer statistics, 2009. CA Cancer J Clin 2009;59:225–49.
- Marugame T, Yoshimi I. Comparison of cancer mortality (lung cancer) in five countries: France, Italy, Japan, UK and USA from the WHO Mortality Database (1960–2000). Jpn J Clin Oncol 2005;35:168–70.
- Marugame T, Mizuno S. Mortality trend of lung cancer in Japan: 1960–2000. Jpn J Clin Oncol 2003;33:148–9.
- Kiyohara C, Otsu A, Shirakawa T, Genetic polymorphisms and lung cancer susceptibility: a review. Lung Cancer 2002;37:241–56.
- Thorgeirsson TE, Geller F, Sulem P, A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008;452:638–42.
- Hung RJ, McKay JD, Gaborieau V, A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008;452:633–7.
- Amos CI, Wu X, Broderick P, Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 2008;40:616–22.
- Shiraishi K, Kohno T, Kunitoh H, Contribution of nicotine acetylcholine receptor polymorphisms to lung cancer risk in a smoking-independent manner in the Japanese. Carcinogenesis 2009;30:65–70.
- Itzkowitz SH, Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol 2004;287:G7–17.
- de Torres JP, Bastarrika G, Wisnivesky JP, Assessing the relationship between lung cancer risk and emphysema detected on low-dose CT of the chest. Chest 2007;132:1932–8.
- Wilson DO, Weissfeld JL, Balkan A, Association of radiographic emphysema and airflow obstruction with lung cancer. Am J Respir Crit Care Med 2008;178:738–44.
- Matsushita H, Tanaka S, Saiki Y, Lung cancer associated with usual interstitial pneumonia. Pathol Int 1995;45:925–32.
- Parimon T, Chien JW, Bryson CL, Inhaled corticosteroids and risk of lung cancer among patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2007;175:712-19.
- Yoshida T, Tuder RM. Pathobiology of cigarette smoke-induced chronic obstructive pulmonary disease. Physiol Rev 2007;87:1047–82.
- McCormack FX, Whitsett JA. The pulmonary collectins, SP-A and SP-D, orchestrate innate immunity in the lung. J Clin Invest 2002;109:707–12.
- Wang Y, Kuan PJ, Xing C, Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am J Hum Genet 2009;84:52–9.
- Ewis AA, Kondo K, Dang F, Surfactant protein B gene variations and susceptibility to lung cancer in chromate workers. Am J Ind Med 2006;49:367–73.
- Sin DD, Man SF, McWilliams A, Surfactant protein D and bronchial dysplasia in smokers at high risk of lung cancer. Chest 2008;134:582–8.
- Ishii T, Hagiwara K, Kamio K, Involvement of surfactant protein D in emphysema revealed by genetic association study. Eur J Hum Genet 2012;20:230–5.
- Foreman MG, Kong X, Demeo DL, Polymorphisms in surfactant protein-D are associated with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol 2011;44:316–22.
- National Heart, Lung, and Blood Institute. The definition of emphysema. Report of a National Heart, Lung, and Blood Institute, Division of Lung Diseases workshop. Am Rev Respir Dis 1985;132:182–5.
- Yamanaka A. Morphopathology of chronic pulmonary emphysema. Acta Pathol Jpn 1965;15:33–9.
- Fujimoto K, Ikeda S, Arai T, Polymorphism of SERPINE2 gene is associated with pulmonary emphysema in consecutive autopsy cases. BMC Med Genet 2010;11:159–65.
- Wright JL, Thurlbeck WM, editors. Emphysema: classification, morphology, and associations. Thurlbeck's Chronic Airflow Obstruction. 2nd ed. Hamilton (ON): B.C. Decker, Inc.; 1999. Chapter 4; p. 85–144.
- Myers JL. Thurlbeck's pathology of the lung. 3rd ed. New York: Thieme Medical Publishers, Inc.; 2005. Chapter 20, Idiopathic interstitial pneumonias; p. 563–600.
- Myers JL. Other diffuse lung diseases. Thurlbeck's Pathology of the Lung. 3rd ed. Wright JL, Thurlbeck WM, editors. New York: Thieme Medical Publishers, Inc., 2005; Chapter 21; pp. 601–74.
- Holland PM, Abramson RD, Watson R, Detection of specific polymerase chain reaction product by utilizing the 52–32 exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA 1991;88:7276–80.
- Baron RM, Kenny DA. The moderator-mediator variable distinction in social psychological research: conceptual, strategic, and statistical considerations. J Pers Soc Psychol 1986;51:1173–82.
- Wang J, Spitz MR, Amos CI, Mediating effects of smoking and chronic obstructive pulmonary disease on the relation between the CHRNA5-A3 genetic locus and lung cancer risk. Cancer 2010;116:3458–62.
- Kenny DA, Kashy DA, Bolger N. Data analysis in social psycho-logy. The Handbook of Social Psychology. Gilbert DT, Fiske ST, Lindzey G, editors. Boston: McGraw-Hill; 1998; p. 233–65.
- MacKinnon A. Estimating mediated effects in prevention studies. Eval Rev 1993;17:144–58.
- Preacher KJ, Hayes AF. SPSS and SAS procedures for estimating indirect effects in simple mediation models. Behav Res Methods Instrum Comput 2004;36:717–31.
- Raghu G, Collard HR, Egan JJ, ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med 2011;183:788–824.
- Leth-Larsen R, Garred P, Jensenius H, A common polymorphism in the SFTPD gene influences assembly, function, and concentration of surfactant protein D. J Immunol 2005;174:1532–8.
- Sorensen GL, Hoegh SV, Leth-Larsen R, Multimeric and trimeric subunit SP-D are interconvertible structures with distinct ligand interaction. Mol Immunol 2009;46:3060–9.
- Shiels MS, Chaturvedi AK, Katki HA, Circulating markers of interstitial lung disease and subsequent risk of lung cancer. Cancer Epidemiol Biomarkers Prev 2011;20:2262–72.
- de Gracia J, Mata F, Alvarez A, Genotype-phenotype correlation for pulmonary function in cystic fibrosis. Thorax 2005;60:558–63.
- Cohen JC, Kiss RS, Pertsemlidis A, Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 2004;305:869–72.
- Matakidou A, Eisen T, Houlston RS. TP53 polymorphisms and lung cancer risk: a systematic review and meta-analysis. Mutagenesis 2003;18:377–85.
- Ligtenberg AJ, Veerman EC, Nieuw Amerongen AV, Salivary agglutinin/glycoprotein-340/DMBT1: a single molecule with variable composition and with different functions in infection, inflammation and cancer. Biol Chem 2007;388:1275–89.
- Wu W, Kemp BL, Proctor ML, Expression of DMBT1, a candidate tumor suppressor gene, is frequently lost in lung cancer. Cancer Res 1999;59:1846–51.
- Bridges JP, Wert SE, Nogee LM, Expression of a human surfactant protein C mutation associated with interstitial lung disease disrupts lung development in transgenic mice. J Biol Chem 2003;278:52739–46.
- Mulugeta S, Nguyen V, Russo SJ, A surfactant protein C precursor protein BRICHOS domain mutation causes endoplasmic reticulum stress, proteasome dysfunction, and caspase 3 activation. Am J Respir Cell Mol Biol 2005;32:521–30.
Appendix (Online Supplemental Material):
Table S1. Genotypes of SFTPD in the subjects with IP, emphysema, and lung cancer